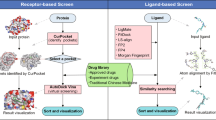
Abstract
The rapidly advancing researches on traditional Chinese medicine (TCM) have greatly intrigued pharmaceutical industries worldwide. To take initiative in the next generation of drug development, we constructed a cloud-computing system for TCM intelligent screening system (iScreen) based on TCM Database@Taiwan. iScreen is compacted web server for TCM docking and followed by customized de novo drug design. We further implemented a protein preparation tool that both extract protein of interest from a raw input file and estimate the size of ligand bind site. In addition, iScreen is designed in user-friendly graphic interface for users who have less experience with the command line systems. For customized docking, multiple docking services, including standard, in-water, pH environment, and flexible docking modes are implemented. Users can download first 200 TCM compounds of best docking results. For TCM de novo drug design, iScreen provides multiple molecular descriptors for a user’s interest. iScreen is the world’s first web server that employs world’s largest TCM database for virtual screening and de novo drug design. We believe our web server can lead TCM research to a new era of drug development. The TCM docking and screening server is available at http://iScreen.cmu.edu.tw/.





Similar content being viewed by others
References
Chen CY, Chen CYC (2010) Insights into designing the dual-targeted HER2/HSP90 inhibitors. J Mol Graphics Model 29(1):21–31
Chang TT, Huang HJ, Lee KJ, Yu HW, Chen HY, Tsai FJ, Sun MF, Chen CY (2010) Key features for designing phosphodiesterase-5 inhibitors. J Biomol Struct Dyn 28(3):309–321
Chen CY, Huang HJ, Tsai FJ, Chen CYC (2010) Drug design for Influenza A virus subtype H1N1. J Taiwan Inst Chem Eng 41(1):8–15
Chen CYC (2010) Virtual screening and drug design for PDE-5 receptor from traditional chinese medicine database. J Biomol Struct Dyn 27(5):627–640
Chen CYC (2009) Chemoinformatics and pharmacoinformatics approach for exploring the GABA-A agonist from Chinese herb suanzaoren. J Taiwan Inst Chem Eng 40(1):36–47
Chen CYC (2009) Computational screening and design of traditional Chinese medicine (TCM) to block phosphodiesterase-5. J Mol Graphics Model 28(4):261–269
Chen KC, Chen CYC (2011) Stroke Prevention by traditional Chinese medicine? A genetic algorithm, support vector machine and molecular dynamics approach. Soft Matter 7(8):4001–4008
Chen CYC (2011) TCM Database@Taiwan: the world’s largest traditional chinese medicine database for drug screening in silico. PLoS One 6(1):e15939
Korb O, Stutzle T, Exner TE (2009) Empirical scoring functions for advanced protein-ligand docking with PLANTS. J Chem Inf Model 49(1):84–96
Jones G, Willett P, Glen RC (1995) Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol 245(1):43–53
Jones G, Willett P, Glen RC, Leach AR, Taylor R (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267(3):727–748
Irwin JJ, Shoichet BK, Mysinger MM, Huang N, Colizzi F, Wassam P, Cao Y (2009) Automated docking screens: a feasibility study. J Med Chem 52(18):5712–5720
Douguet D, Munier-Lehmann H, Labesse G, Pochet S (2005) LEA3D: a computer-aided ligand design for structure-based drug design. J Med Chem 48(7):2457–2468
Wass MN, Kelley LA, Sternberg MJ (2010) 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res 38(Web Server issue):W469–W473
Liu X, Ouyang S, Yu B, Liu Y, Huang K, Gong J, Zheng S, Li Z, Li H, Jiang H (2010) PharmMapper server: a web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res 38(Web Server issue):W609–W614
Dey F, Caflisch A (2008) Fragment-based de novo ligand design by multiobjective evolutionary optimization. J Chem Inf Model 48(3):679–690
Law JMS, Fung DYK, Zsoldos Z, Simon A, Szabo Z, Csizmadia IG, Johnson AP (2003) Validation of the SPROUT de novo design program. Theochem-J Mol Struct 666:651–657
Danziger DJ, Dean PM (1989) Automated site-directed drug design: the prediction and observation of ligand point positions at hydrogen-bonding regions on protein surfaces. Proc R Soc Lond B Biol Sci 236(1283):115–124
Nishibata Y, Itai A (1993) Confirmation of usefulness of a structure construction program based on three-dimensional receptor structure for rational lead generation. J Med Chem 36(20):2921–2928
Murray CW, Clark DE, Byrne DG (1995) PRO_LIGAND: an approach to de novo molecular design. 6. Flexible fitting in the design of peptides. J Comput-Aided Mol Des 9(5):381–395
Rotstein SH, Murcko MA (1993) GenStar: a method for de novo drug design. J Comput-Aided Mol Des 7(1):23–43
Bohm HJ (1992) The computer program LUDI: a new method for the de novo design of enzyme inhibitors. J Comput-Aided Mol Des 6(1):61–78
Roe DC, Kuntz ID (1995) BUILDER v.2: improving the chemistry of a de novo design strategy. J Comput-Aided Mol Des 9(3):269–282
Pearlman DA, Murcko MA (1996) CONCERTS: dynamic connection of fragments as an approach to de novo ligand design. J Med Chem 39(8):1651–1663
Vinkers HM, de Jonge MR, Daeyaert FF, Heeres J, Koymans LM, van Lenthe JH, Lewi PJ, Timmerman H, Van Aken K, Janssen PA (2003) SYNOPSIS: synthesize and optimize system in silico. J Med Chem 46(13):2765–2773
Brown N, McKay B, Gilardoni F, Gasteiger J (2004) A graph-based genetic algorithm and its application to the multiobjective evolution of median molecules. J Chem Inf Comput Sci 44(3):1079–1087
Chen G, Li S (1992) Ben cao gang mu tong shi = general explanation of compendium of materia medica. Xue yuan chu ban she, Beijing Shi
Fang Y, Zhang Z, Miao X (1991) Shang han lun tiao bian, Shanghai gu ji chu ban she : Xin hua shu dian Shanghai fa xing suo fa xing, Shanghai
Lü X (2002) Zhong yao jian bie da quan. Hunan ke xue ji shu chu ban she, Changsha Shi
Miao X, Zheng J (2002) Shennong ben cao jing shu. Zhong yi gu ji chu ban she, Beijing
Nanjing Zhong yi yao da xue, Zhao G, Dai S, Chen R (2006) Zhong yao da ci dian. Shanghai ke xue ji shu chu ban she, Shanghai
Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Lu ZY, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Mizrachi I, Ostell J, Panchenko A, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Slotta D, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Wang YL, Wilbur WJ, Yaschenko E, Ye J (2010) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 38:D5–D16
Clark M (1998) The ISI web of science. Abstracts of Papers of the American Chemical Society 216:U525
Gunda T (2007) ChemBioOffice. Chem World 4(11):70
Karzazi Y, Surpateanu G (1999) An empirical MM2 augmented force field for the cycloimmonium ylides. J Mol Struct 510(1–3):197–205
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Del Rev 46(1–3):3–26
Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A (2005) H ++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res 33(Web Server issue):W368–W371
Acknowledgments
The research was supported by grants from the National Science Council of Taiwan (NSC 99-2221-E-039-013-), Committee on Chinese Medicine and Pharmacy (CCMP100-RD-030) China Medical University and Asia University (CMU99-TCM, CMU99-S-02, CMU99-ASIA-25, CMU99-ASIA-26 CMU99-ASIA-27 CMU99-ASIA-28). This study is also supported in part by Taiwan Department of Health Clinical Trial and Research Center of Excellence (DOH100-TD-B-111-004) and Taiwan Department of Health Cancer Research Center of Excellence (DOH100-TD-C-111-005). We are grateful to the Asia University cloud-computing facilities.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tsai, TY., Chang, KW. & Chen, C.YC. iScreen: world’s first cloud-computing web server for virtual screening and de novo drug design based on TCM database@Taiwan . J Comput Aided Mol Des 25, 525–531 (2011). https://doi.org/10.1007/s10822-011-9438-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-011-9438-9