Abstract
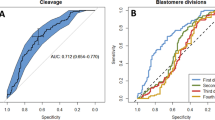
To assess whether morphokinetic features at the cleavage stage together with specific gene expression in cumulus cells (CCs) may be used to predict whether human embryos are able to achieve the expanded blastocyst stage on day 5. Eighty-one embryos were cultured using the Geri plus® time-lapse system. Twenty-seven embryos progressing to the expanded blastocyst stage (BL group) were compared with thirty-five embryos showing developmental arrest (AR group) and nineteen reaching the stage of early or not fully expanded blastocyst (nBL group). The analyzed morphokinetic variables were pronuclear appearance (tPNa), pronuclear fading (tPNf), and completion of cleavage to two, three, four, and eight cells (t2, t3, t4, and t8). CCs were analyzed by RT-qPCR for bone morphogenetic protein 15 (BMP15), cytochrome c oxidase subunit II (COXII), ATP synthase subunit 6 (MT-ATP6), connexin 43 (Cx43), and heme oxygenase-1 (HO-1). Embryos of BL group showed a significantly faster kinetic. BMP15, COXII, and MT-ATP6 mRNA expression was significantly higher in CCs of BL group embryos, whereas Cx43 and HO-1 mRNA levels were higher in AR group. Kinetic parameters and gene expression were not significantly different between either the BL and nBL groups or the AR and nBL groups. ROC curves showed that the most predictive cut-offs were t2 < 26.25 for morphokinetics and COXII > 0.3 for gene expression. Multivariable logistic regression analysis showed that morphokinetic variables and gene expression were both valuable, independent predictors of embryo development to expanded blastocyst. Our results suggest the possibility of developing integrated prediction models for early embryo selection at the cleavage stage.




Similar content being viewed by others
References
Glujovsky D, Farquhar C, Quinteiro Retamar AM, Alvarez Sedo CR, Blake D. Cleavage stage versus blastocyst stage embryo transfer in assisted reproductive technology. Cochrane Database Syst Rev. 2016;CD002118.
White CR, Denomme MM, Tekpetey FR, Feyles V, Power SGA, Mann MRW. High frequency of imprinted methylation errors in human preimplantation embryos. Sci Rep. 2015;5:17311.
Gardner DK, Balaban B. Assessment of human embryo development using morphological criteria in an era of time-lapse, algorithms and “OMICS”: is looking good still important? Mol Hum Reprod. 2016;22:704–18.
Meseguer M, Herrero J, Tejera A, Hilligsøe KM, Ramsing NB, Remohí J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod Oxf Engl. 2011;26:2658–71.
Conaghan J, Chen AA, Willman SP, Ivani K, Chenette PE, Boostanfar R, et al. Improving embryo selection using a computer-automated time-lapse image analysis test plus day 3 morphology: results from a prospective multicenter trial. Fertil Steril. 2013;100:412–419.e5.
Diamond MP, Suraj V, Behnke EJ, Yang X, Angle MJ, Lambe-Steinmiller JC, et al. Using the Eeva TestTM adjunctively to traditional day 3 morphology is informative for consistent embryo assessment within a panel of embryologists with diverse experience. J Assist Reprod Genet. 2015;32:61–8.
Kieslinger DC, De Gheselle S, Lambalk CB, De Sutter P, Kostelijk EH, Twisk JWR, et al. Embryo selection using time-lapse analysis (Early Embryo Viability Assessment) in conjunction with standard morphology: a prospective two-center pilot study. Hum Reprod Oxf Engl. 2016;31:2450–7.
Fragouli E, Lalioti MD, Wells D. The transcriptome of follicular cells: biological insights and clinical implications for the treatment of infertility. Hum Reprod Update. 2014;20:1–11.
Huang Z, Wells D. The human oocyte and cumulus cells relationship: new insights from the cumulus cell transcriptome. Mol Hum Reprod. 2010;16:715–25.
Demiray SB, Goker ENT, Tavmergen E, Yilmaz O, Calimlioglu N, Soykam HO, et al. Differential gene expression analysis of human cumulus cells. Clin Exp Reprod Med. 2019;46:76–86.
Li Y, Li R-Q, Ou S-B, Zhang N-F, Ren L, Wei L-N, et al. Increased GDF9 and BMP15 mRNA levels in cumulus granulosa cells correlate with oocyte maturation, fertilization, and embryo quality in humans. Reprod Biol Endocrinol RBE. 2014;12:81.
Persani L, Rossetti R, Di Pasquale E, Cacciatore C, Fabre S. The fundamental role of bone morphogenetic protein 15 in ovarian function and its involvement in female fertility disorders. Hum Reprod Update. 2014;20:869–83.
Chang H-M, Qiao J, Leung PCK. Oocyte-somatic cell interactions in the human ovary-novel role of bone morphogenetic proteins and growth differentiation factors. Hum Reprod Update. 2016;23:1–18.
Sanfins A, Rodrigues P, Albertini DF. GDF-9 and BMP-15 direct the follicle symphony. J Assist Reprod Genet. 2018;35:1741–50.
Winterhager E, Kidder GM. Gap junction connexins in female reproductive organs: implications for women’s reproductive health. Hum Reprod Update. 2015;21:340–52.
Donfack NJ, Alves KA, Alves BG, Rocha RMP, Bruno JB, Bertolini M, et al. Stroma cell-derived factor 1 and connexins (37 and 43) are preserved after vitrification and in vitro culture of goat ovarian cortex. Theriogenology. 2018;116:83–8.
Bergandi L, Basso G, Evangelista F, Canosa S, Dalmasso P, Aldieri E, et al. Inducible nitric oxide synthase and heme oxygenase 1 are expressed in human cumulus cells and may be used as biomarkers of oocyte competence. Reprod Sci Thousand Oaks Calif. 2014;21:1370–7.
Revelli A, Canosa S, Bergandi L, Skorokhod OA, Biasoni V, Carosso A, et al. Oocyte polarized light microscopy, assay of specific follicular fluid metabolites, and gene expression in cumulus cells as different approaches to predict fertilization efficiency after ICSI. Reprod Biol Endocrinol RBE. 2017;15:47.
Huber M, Hadziosmanovic N, Berglund L, Holte J. Using the ovarian sensitivity index to define poor, normal, and high response after controlled ovarian hyperstimulation in the long gonadotropin-releasing hormone-agonist protocol: suggestions for a new principle to solve an old problem. Fertil Steril. 2013;100:1270–6.
Canosa S, Adriaenssens T, Coucke W, Dalmasso P, Revelli A, Benedetto C, et al. Zona pellucida gene mRNA expression in human oocytes is related to oocyte maturity, zona inner layer retardance and fertilization competence. Mol Hum Reprod. 2017;23:292–303.
Holte J, Berglund L, Milton K, Garello C, Gennarelli G, Revelli A, et al. Construction of an evidence-based integrated morphology cleavage embryo score for implantation potential of embryos scored and transferred on day 2 after oocyte retrieval. Hum Reprod Oxf Engl. 2007;22:548–57.
Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology. The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod Oxf Engl. 2011;26:1270–83.
Revelli A, Rovei V, Dalmasso P, Gennarelli G, Racca C, Evangelista F, et al. Large randomized trial comparing transabdominal ultrasound-guided embryo transfer with a technique based on uterine length measurement before embryo transfer. Ultrasound Obstet Gynecol. 2016;48(3):289–95.
Bayne RAL, Kinnell HL, Coutts SM, He J, Childs AJ, Anderson RA. GDF9 is transiently expressed in oocytes before follicle formation in the human fetal ovary and is regulated by a novel NOBOX transcript. PloS One. 2015;10:e0119819.
Chang H-M, Cheng J-C, Taylor E, Leung PCK. Oocyte-derived BMP15 but not GDF9 down-regulates connexin43 expression and decreases gap junction intercellular communication activity in immortalized human granulosa cells. Mol Hum Reprod. 2014;20:373–83.
Ricca C, Aillon A, Bergandi L, Alotto D, Castagnoli C, Silvagno F. Vitamin D receptor is necessary for mitochondrial function and cell health. Int J Mol Sci. 2018;19.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034.
Pribenszky C, Nilselid A-M, Montag M. Time-lapse culture with morphokinetic embryo selection improves pregnancy and live birth chances and reduces early pregnancy loss: a meta-analysis. Reprod Biomed Online. 2017;35:511–20.
Armstrong S, Arroll N, Cree LM, Jordan V, Farquhar C. Time-lapse systems for embryo incubation and assessment in assisted reproduction. Cochrane Database Syst Rev. 2015;CD011320.
Armstrong S, Vail A, Mastenbroek S, Jordan V, Farquhar C. Time-lapse in the IVF-lab: how should we assess potential benefit? Hum Reprod Oxf Engl. 2015;30:3–8.
Chen M, Wei S, Hu J, Yuan J, Liu F. Does time-lapse imaging have favorable results for embryo incubation and selection compared with conventional methods in clinical in vitro fertilization? A meta-analysis and systematic review of randomized controlled trials. PloS One. 2017;12:e0178720.
Polanski LT, Coelho Neto MA, Nastri CO, Navarro PA, Ferriani RA, Raine-Fenning N, et al. Time-lapse embryo imaging for improving reproductive outcomes: systematic review and meta-analysis. Ultrasound Obstet Gynecol Off J Int Soc Ultrasound Obstet Gynecol. 2014;44:394–401.
Racowsky C, Kovacs P, Martins WP. A critical appraisal of time-lapse imaging for embryo selection: where are we and where do we need to go? J Assist Reprod Genet. 2015;32:1025–30.
Khosravi P, Kazemi E, Zhan Q, Malmsten JE, Toschi M, Zisimopoulos P, et al. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. Npj Digit Med. 2019;2:21.
Coticchio G, Mignini Renzini M, Novara PV, Lain M, De Ponti E, Turchi D, et al. Focused time-lapse analysis reveals novel aspects of human fertilization and suggests new parameters of embryo viability. Hum Reprod Oxf Engl. 2018;33:23–31.
Revelli A, Canosa S, Carosso A, Filippini C, Paschero C, Gennarelli G, et al. Impact of the addition of Early Embryo Viability Assessment to morphological evaluation on the accuracy of embryo selection on day 3 or day 5: a retrospective analysis. J Ovarian Res. 2019;12:73.
Zaninovic N, Irani M, Meseguer M. Assessment of embryo morphology and developmental dynamics by time-lapse microscopy: is there a relation to implantation and ploidy? Fertil Steril. 2017;108:722–9.
Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM, et al. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol. 2010;28:1115–21.
Hammond ER, Stewart B, Peek JC, Shelling AN, Cree LM. Assessing embryo quality by combining non-invasive markers: early time-lapse parameters reflect gene expression in associated cumulus cells. Hum Reprod Oxf Engl. 2015;30:1850–60.
Hashimoto S, Kato N, Saeki K, Morimoto Y. Selection of high-potential embryos by culture in poly(dimethylsiloxane) microwells and time-lapse imaging. Fertil Steril. 2012;97:332–7.
Kirkegaard K, Kesmodel US, Hindkjær JJ, Ingerslev HJ. Time-lapse parameters as predictors of blastocyst development and pregnancy outcome in embryos from good prognosis patients: a prospective cohort study. Hum Reprod. 2013;28:2643–51.
Cruz M, Garrido N, Herrero J, Pérez-Cano I, Muñoz M, Meseguer M. Timing of cell division in human cleavage-stage embryos is linked with blastocyst formation and quality. Reprod Biomed Online. 2012;25:371–81.
Dal Canto M, Coticchio G, Mignini Renzini M, De Ponti E, Novara PV, Brambillasca F, et al. Cleavage kinetics analysis of human embryos predicts development to blastocyst and implantation. Reprod Biomed Online. 2012;25:474–80.
Minasi MG, Colasante A, Riccio T, Ruberti A, Casciani V, Scarselli F, et al. Correlation between aneuploidy, standard morphology evaluation and morphokinetic development in 1730 biopsied blastocysts: a consecutive case series study. Hum Reprod Oxf Engl. 2016;31:2245–54.
Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Hickman CFL. Modelling a risk classification of aneuploidy in human embryos using non-invasive morphokinetics. Reprod Biomed Online. 2013;26:477–85.
Shapiro BS, Richter KS, Harris DC, Daneshmand ST. A comparison of day 5 and day 6 blastocyst transfers. Fertil Steril. 2001;75:1126–30.
Barrenetxea G, López de Larruzea A, Ganzabal T, Jiménez R, Carbonero K, Mandiola M. Blastocyst culture after repeated failure of cleavage-stage embryo transfers: a comparison of day 5 and day 6 transfers. Fertil Steril. 2005;83:49–53.
Franasiak JM, Forman EJ, Patounakis G, Hong KH, Werner MD, Upham KM, et al. Investigating the impact of the timing of blastulation on implantation: management of embryo-endometrial synchrony improves outcomes. Hum Reprod Open. 2018;2018:hoy022.
Tannus S, Cohen Y, Henderson S, Al Ma’mari N, Shavit T, Son W-Y, et al. Fresh transfer of day 5 slow-growing embryos versus deferred transfer of vitrified, fully expanded day 6 blastocysts: which is the optimal approach? Hum Reprod Oxf Engl. 2019;34:44–51.
Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van de Velde H, Coucke W, et al. Cumulus cell gene expression predicts better cleavage-stage embryo or blastocyst development and pregnancy for ICSI patients. Hum Reprod Oxf Engl. 2011;26:1035–51.
Wathlet S, Adriaenssens T, Segers I, Verheyen G, Janssens R, Coucke W, et al. New candidate genes to predict pregnancy outcome in single embryo transfer cycles when using cumulus cell gene expression. Fertil Steril. 2012;98:432–439.e1-4.
Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van Landuyt L, Coucke W, et al. Pregnancy prediction in single embryo transfer cycles after ICSI using QPCR: validation in oocytes from the same cohort. PloS One. 2013;8:e54226.
Wu Y-T, Tang L, Cai J, Lu X-E, Xu J, Zhu X-M, et al. High bone morphogenetic protein-15 level in follicular fluid is associated with high quality oocyte and subsequent embryonic development. Hum Reprod Oxf Engl. 2007;22:1526–31.
Hasegawa J, Yanaihara A, Iwasaki S, Mitsukawa K, Negishi M, Okai T. Reduction of connexin 43 in human cumulus cells yields good embryo competence during ICSI. J Assist Reprod Genet. 2007;24:463–6.
Sutton-McDowall ML, Mottershead DG, Gardner DK, Gilchrist RB, Thompson JG. Metabolic differences in bovine cumulus-oocyte complexes matured in vitro in the presence or absence of follicle-stimulating hormone and bone morphogenetic protein 15. Biol Reprod. 2012;87:87.
Ricca C, Aillon A, Viano M, Bergandi L, Aldieri E, Silvagno F. Vitamin D inhibits the epithelial-mesenchymal transition by a negative feedback regulation of TGF-β activity. J Steroid Biochem Mol Biol. 2019;187:97–105.
Danyu L, Yanran L, Xiuna J, Ying C, Sudan P, Tianen Z, et al. α-Synuclein induced mitochondrial dysfunction via cytochrome c oxidase subunit 2 in SH-SY5Y cells. Exp Cell Res. 2019;378:57–65.
Ganetzky RD, Stendel C, McCormick EM, Zolkipli-Cunningham Z, Goldstein AC, Klopstock T, et al. MT-ATP6 mitochondrial disease variants: phenotypic and biochemical features analysis in 218 published cases and cohort of 14 new cases. Hum Mutat. 2019;40:499–515.
Bansal S, Biswas G, Avadhani NG. Mitochondria-targeted heme oxygenase-1 induces oxidative stress and mitochondrial dysfunction in macrophages, kidney fibroblasts and in chronic alcohol hepatotoxicity. Redox Biol. 2014;2:273–83.
Scarica C, Cimadomo D, Dovere L, Giancani A, Stoppa M, Capalbo A, et al. An integrated investigation of oocyte developmental competence: expression of key genes in human cumulus cells, morphokinetics of early divisions, blastulation, and euploidy. J Assist Reprod Genet. 2019;36:875–87.
Acknowledgments
This work was supported by MIUR in the PhD (for S.C.) and Postdoc Program (for L.B.) of the University of Torino, Italy.
Author information
Authors and Affiliations
Contributions
A.R., S.C., and L.B. conceived the study; A.C. and N.D.S. enrolled the patients; S.C., C.M., and C.P. were responsible for the time-lapse data acquisition and analysis; L.B. and C.M. performed real-time polymerase chain reaction experiments; L.C. performed the statistical analysis and S.C. contributed to the statistical analysis; S.C. and L.B. contributed to the validation, the interpretation of the results, the writing and editing of the manuscript; F.S., G.G., C.B., and A.R. contributed to the final interpretation of the data and editing of the manuscript. All authors gave their final approval.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Stefano Canosa and Loredana Bergandi are coauthors
Rights and permissions
About this article
Cite this article
Canosa, S., Bergandi, L., Macrì, C. et al. Morphokinetic analysis of cleavage stage embryos and assessment of specific gene expression in cumulus cells independently predict human embryo development to expanded blastocyst: a preliminary study. J Assist Reprod Genet 37, 1409–1420 (2020). https://doi.org/10.1007/s10815-020-01806-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-020-01806-6