Abstract
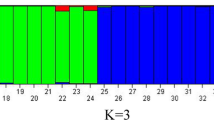
Cicer arietinum L. (chickpea) is one of the most significant legume crops domesticated in the Fertile Crescent. This study was aimed to characterize a diverse composite set of 384 Cicer genotypes using unlinked simple sequence repeat (SSR) markers. The genotypes grown under the temperate conditions of Western-Himalayas included cultivated and wild relatives from primary (Cicer reticulatum Ladiz.), secondary (Cicer echinospermum P.H. Davis) and tertiary (Cicer microphyllum Benth.) gene pools. The analysis of genotypic data of eight SSR markers from eight linkage groups led to the identification of 63 alleles, ranging from 2 to 6 with an average value of 3.7 alleles per locus. The polymorphic information content of SSR markers ranged from 0.46 to 0.79 with an average value of 0.77 and the gene diversity ranged from 0.47 to 0.79 with an average of 0.64. The clustering of genotypes in the form of dendrogram discriminated all 384 genotypes into four major clusters. The wild genotypes belonging to different gene pools got clustered uniformly in different clusters along with cultivated chickpea genotypes. The analysis of data also led to the selection of core set of 192 genotypes. The core set was found to possess same diversity (63 alleles; average alleles per locus: 3.7; gene diversity: 0.65) as that of composite set of 384 genotypes. The development of core set in chickpea shall prove useful in gene discovery for variety of traits through genome-wide association studies. The results also provide an insight into gene/allele diversity available in our chickpea germplasm collection grown under agro-climatic conditions of the North Western-Himalayas.




Similar content being viewed by others
Data availability
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.
References
Brown AHD, Frankel OH, Marshall DR, Williams JT (1989) The use of plant genetic resources. Cambridge University Press. Cambridge, UK, pp 136–155
Cevik S, Unyayar S, Ergul A (2015) Genetic relationships between cultivars of Cicer arietinum and its progenitor grown in Turkey determined by using the SSR markers. Turk J Field Crops 20(1):109–114
Choudhary P, Khanna SM, Jain PK, Bharadwaj C, Kumar J, Lakhera PC, Srinivasan R (2012) Genetic structure and diversity analysis of the primary gene pool of chickpea using SSR markers. Genet Mol Res 11:891–905
Choudhary P, Khanna SM, Jain PK, Bharadwaj C, Kumar J, Lakhera PC, Srinivasan R (2013) Molecular characterization of primary gene pool of chickpea based on ISSR markers. Biochem Genet 51:306–322
Choudhury DR, Singh N, Singh AK, Kumar S, Srinivasan K, Tyagi RK, Singh R (2014) Analysis of genetic diversity and population structure of rice germplasm from North-Eastern region of India and development of a core germplasm set. PLoS One 9(11):e113094
Díez CM, Imperato A, Rallo L, Barranco D, Trujillo I (2012) Worldwide core collection of olive cultivars based on simple sequence repeat and morphological markers. Crop Sci 52(1):211–221
Gaur PM, Jukanti AK, Varshney RK (2012) Impact of genomic technologies on chickpea breeding strategies. Agronomy 2:199–221
Ghaffari P, Talebi R, Keshavarz F (2014) Genetic diversity and geographical differentiation of Iranian landrace, cultivars and exotic chickpea lines as revealed by morphological and microsatellite markers. Physiol Mol Biol Plant 20:225–233
Gupta PK, Varshney RK (2000) The development and use of microsatellite markers for genetics and plant breeding with emphasis on bread wheat. Euphytica 1(13):163–185
Gupta PK, Rustgi S, Mir RR (2008) Array-based high-throughput DNA markers for crop improvement. Heredity 101:5–18
PK Gupta, S Rustgi, RR Mir (2013) Array-based high-throughput DNA markers and genotyping platforms for cereal genetics and genomics. In: P.K Gupta and R.K Varshney (Eds.), Cereal Genomics II. Springer: The Netherlands
Hajibarat Z, Saidi A, Hajibarat Z, Talebi R (2014) Genetic diversity and population structure analysis of landrace and improved chickpea (Cicer arietinum) genotypes using morphological and microsatellite markers. Environ Expt Biol 12:161–166
Hajibarat Z, Saidi A, Hajibarat Z, Talebi R (2015) Characterization of genetic diversity in chickpea using SSR markers, start codon targeted polymorphism (SCoT) and conserved DNA-derived polymorphism (CDDP). Physiol Mol Biol Plants 21:365–373
Hüttel B, Winter P, Weising K (1999) Sequence tagged microsatellite site markers for chickpea (Cicer arietinum L.). Genome 42:210–217
Fayaz H, Rather IA, Wani AA, Tyagi S, Pandey R, Mir RR (2019) Characterization of chickpea gene pools for nutrient concentrations under agro-climatic conditions of North-Western Himalayas. Plant Genet Resour Charact Util 17:464–467
Iruela M, Rubio J, Cubero JI, Gil J, Milan T (2002) Phylogenetic analysis in the genus Cicer and cultivated chickpea using RAPD and ISSR markers. Theor Appl Genet 104:643–651
Jha UC, Jha R, Bohra A, Parida SK, Kole PC, Thakro V, Singh D, Singh NP (2018) Population structure and association analysis of heat stress relevant traits in chickpea (Cicer arietinum L.). Biotech 8(1):43
Keneni G, Bekele E, Imtiaz M, Dagne K, Getu E, Assefa F (2012) Genetic diversity and population structure of Ethiopian chickpea (Cicer arietinum L.) germplasm accessions from different geographical origins as revealed by microsatellite markers. Plant Mol Biol Rep 30:654–665
Kumar, S., Kumar, M., Mir, R. R., Kumar, R., Kumar, S. (2021) Advances in Molecular Markers and Their Use in Genetic Improvement of Wheat. In Wani SH, Mohan A and Singh GP (Eds.). Physiological, Molecular, and Genetic Perspectives of Wheat Improvement pp 139–174. https://doi.org/10.1007/978-3-030-59577-7_8
Mallikarjuna BP, Samineni S, Thudi M, Sajja SB, Khan AW, Patil A, Viswanatha KP, Varshney RK, Gaur PM (2017) Molecular mapping of flowering time major genes and QTLs in Chickpea (Cicer arietinum L.). Front Plant Sci 8:1140
Mir RR, Varshney RK (2013a) Future prospects of molecular markers in plants. In: Henry RJ (ed) Molecular markers in plants. Blackwell Publishing Ltd, Oxford, UK., pp 169–190
Mir RR, Hiremath PJ, Riera-Lizarazu O, Varshney RK (2013b) Evolving molecular marker technologies in plants: from RFLPs to GBS. In: Lübberstedt T, Varshney RK (eds) Diagnostics in plant breeding. Springer, New York, pp 229–247
Mir AH, Bhat MA, Dar SA, Sofi PA, Bhat NA, Mir RR (2021) Assessment of cold tolerance in chickpea (Cicer spp.) grown under cold/freezing weather conditions of North-Western Himalayas of Jammu and Kashmir, India. Physiol Mol Biol Plants. https://doi.org/10.1007/s12298-021-00997-1
Naghavi MR, Monfared SR, Humberto G (2012) Genetic diversity in Iranian chickpea (Cicer arietinum L.) landraces as revealed by microsatellite markers. Czech J Genet Plant Breed 48:131–138
Nayak SN, Zhu H, Varghese N (2010) Integration of novel SSR and genebased SNP marker loci in the chickpea genetic map and establishment of new anchor points with Medicago truncatula genome. Theor Appl Genet 120:1415–1441
Nevo E (1978) Genetic variation in natural populations: patterns and theory. Theor Popul Biol 13:121–177
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Perrier X, Jacquemoud-Collet JP (2006) Darwin software. http://darwin.cirad.fr/darwin
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Rao ES, Kadirvel P, Symonds RC, Geethanjali S, Ebert AW (2012) Using SSR markers to map genetic diversity and population structure of Solanum pimpinellifolium for development of a core collection. Plant Genet Resour 10(1):38–48
Saeed A, Hovsepyan H, Darvishzadeh R, Imtiaz M, Panguluri SK, Nazaryan R (2011) Genetic diversity of Iranian accessions, improved lines of chickpea (Cicer arietinum L.) and their wild relatives by using simple sequence repeats. Plant Mol Biol Rep 29:848–858
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer length polymorphism in barley: Mendelian inheritance, chromosomal location and population dynamic. Proc Natl Acad Sci USA 81:8014–8019
Sant VJ, Patankar AG, Sarode ND (1999) Potential of DNA markers in detecting divergence and in analyzing heterosis in Indian elite chickpea cultivars. Theor Appl Genet 98:1217–1225
Sefera T, Abebie B, Gaur PM, Assefa K, Varshney RK (2011) Characterisation and genetic diversity analysis of selected chickpea cultivars of nine countries using simple sequence repeat (SSR) markers. Crop Pasture Sci 62(2):177–187
Singh R, Durga Prasad C, Singhal V, Randhawa GJ (2003) Assessment of genetic diversity in chickpea cultivars using RAPD, AFLP and STMS markers. J Genet Breeding 57:165–174
Singh R, Singhal V, Randhawa GJ (2008) Molecular analysis of chickpea (Cicer arietinum L.) cultivars using AFLP and STMS markers. J Plant Biochem Biotechnol 17:167–171
Sihag P, Sagwal V, Kumar A, Balyan P, Mir RR, Dhankher OP, Kumar O (2021) Discovery of miRNAs and development of heat- responsive miRNA-SSR markers for characterization of wheat germplasm for terminal heat tolerance breeding. Front Genet (plant Genomics). https://doi.org/10.3389/fgene.2021.699420
Sonnante G, Marangi A, Venora G, Pignone D (1997) Using RAPD markers to investigate genetic variation in chickpea. J Genet Breed 51:303–307
Talebi R, Naji AM, Fayaz F (2008a) Geographical patterns of genetic diversity in cultivated chickpea (Cicer arietinum L.) characterized by amplified fragment length polymorphism. Plant Soil Environ 54:447–452
Talebi R, Fayaz R, Mardi M, Pirsyedi SM, Naji AM (2008b) Genetic relationships among chickpea (Cicer arietinum) elite lines based on RAPD and agronomic markers. Int J Agr Biol 8:1560–8530
Tyagi S, Kumar A, Gautam T, Pandey R, Rustgi S, Mir RR (2021) Development and use of miRNA-derived SSR markers for the study of genetic diversity, population structure, and characterization of genotypes for breeding heat tolerant wheat varieties. PLOS One 16(2):e0231063
Tyagi S, Sharma S, Ganie SA, Tahir M, Mir RR, Pandey R (2019) Plant microRNAs: biogenesis, gene silencing, web-based analysis tools and their use as molecular markers. Biotech 9(11):413
Udupa SM, Sharma A, Sharma RP, Pai RA (1993) Narrow genetic variability in Cicer arietinum L. as revealed by RFLP analysis. Plant Biochem Biotech 2:83–86
Upadhyaya HD, Dwivedi SL, Baum M, Varshney RK, Udupa SM, Gowda CLL, Hoisington DA, Singh S (2008) Genetic structure, diversity, and allelic richness in composite collection and reference set in chickpea (Cicer arietinum L.). BMC Plant Biol 8(1):106
Upadhyaya HD, Reddy KN, Sharma S, Varshney RK, Bhattacharjee R, Singh S, Gowda CLL (2011) Pigeonpea composite collection and identification of germplasm for use in crop improvement programmes. Plant Genet Resourc 9(1):97–108
Varshney RK, Hoisington DA, Upadhyaya HD (2007) Molecular genetics and breeding of grain legume crops for the semi-arid tropics. Genomics assisted crop improvement. Springer, Dordrecht, The Netherlands, pp 207–242
Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, Cannon S, Baek J, Rosen BD, Tar’an B, Millan T (2013) Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol 31(3):240
Acknowledgements
The authors are thankful to ICRISAT, Hyderabad, IIPR, Kanpur, NBPGR, New Delhi for providing the germplasm for the present study. Thanks are also due to Douglas R cook at University of California at Davis (UC-Davis), California, USA for providing seeds of some wild relatives through ICRISAT, Hyderabad. The authors are also thankful to the head Division of Genetics and Plant Breeding SKAUT-K for providing the necessary facilities during the course of study. The corresponding author is also thankful to various funding agencies including Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Government of India for providing different grants in the form of NPDFs to students mentored by me and to Department of Biotechnology (DBT), GOI, New Delhi, India.
Funding
This study was not specifically funded by any funding agency.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fayaz, H., Mir, A.H., Tyagi, S. et al. Assessment of molecular genetic diversity of 384 chickpea genotypes and development of core set of 192 genotypes for chickpea improvement programs. Genet Resour Crop Evol 69, 1193–1205 (2022). https://doi.org/10.1007/s10722-021-01296-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01296-0