Abstract
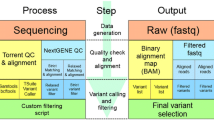
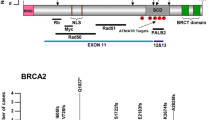
BRCA1 and BRCA2 genes are implicated in 20–25% of hereditary breast and ovarian cancers. New age sequencing platforms have revolutionized massively parallel sequencing in clinical practice by providing cost effective, rapid, and sensitive sequencing. This study critically evaluates the false positives in multiplex panels and suggests the need for careful analysis. We employed multiplex PCR based BRCA1 and BRCA2 community Panel with ion torrent PGM machine for evaluation of these mutations. Out of all 41samples analyzed for BRCA1 and BRCA2 five were found with 950_951 insA(Asn319fs) at Chr13:32906565 position and one sample with 1032_1033 insA(Asn346fs) at Chr13:32906647, both being frame-shift mutations in BRCA2 gene. 950_951 insA(Asn319fs) mutation is reported as pathogenic allele in NCBI dbSNP. On examination of IGV for all these samples, it was seen that both mutations had ‘A’ nucleotide insertion at 950, and 1032 position in exon 10 of BRCA2 gene. Sanger Sequencing did not confirm these insertions. Next-generation sequencing shows great promise by allowing rapid mutational analysis of multiple genes in human cancer but our results indicate the need for careful sequence analysis to avoid false positive results.


Similar content being viewed by others
References
Easton DF (1999) How many more breast cancer predisposition genes are there? Breast Cancer Res 1:14–17
Campeau PM, Foulkes WD, Tischkowitz MD et al (2008) Hereditary breast cancer: new genetic developments, new therapeutic avenues. Hum Genet 124:31–42
Pal T, Permuth-Wey J, Betts JA et al (2005) BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer 04:2807–2816
Petrucelli N, Daly M, Feldman G et al (2016) BRCA1 and BRCA2 hereditary breast and ovarian cancer. Gene Reviews University of Washington, Seattle 1993–2016
Zhang J, Fackenthal JD, Huo D et al (2010) Searching for large genomic rearrangements of the BRCA1 gene in a Nigerian population. Breast Cancer Res Treat 124:573–577
Mafficini A, Simbolo M, Parisi A et al (2016) BRCA somatic and germline mutation detection in paraffin embedded ovarian cancers by next-generation sequencing. Oncotarget 7:1076–1083
Pisano M, Mezzollo V, Galante MM et al (2011) A new mutation of BRCA2 gene in an Italian healthy woman with familial breast cancer history. Fam Cancer 10:65–71
Rothberg JM, Hinz W, Rearick TM et al (2011) An integrated semiconductor device enabling non-optical genome sequencing. Nature 475:348–352
Voelkerding KV, Dames SA, Durtschi JD et al (2009) Next-generation sequencing: from basic research to diagnostics. Clin Chem 55:641–658
Bragg LM, Stone G, Butler KM et al (2013) Shining a light on dark sequencing: characterising errors in Ion Torrent PGM data. PLoS Comput Biol 9:e1003031
Stuewing JP, Abeliovich D, Petertz T et al (1995) The carrier frequency of the BRCA1 mutation 185delAG mutation is approximately 1 percent in Ashkenazi Jewish individuals. Nat Genet 11:198–200
Johannsson O, Ostemeyer EA, Hakansson S et al (1996) Founding BRCA1 mutations in hereditary breast and ovarian cancer in southern Sweden. Am J Hum Genet 58:441–450
Fitzgerald MG, MacDonald DJ, Krainer M et al (1996) Germline BRCA1 mutations in Jewish and non-Jewish women with early onset breast cancer. N Engl J Med 334:143–149
Haliassos A, Chomel JC, Tesson L et al (1989) Modification of enzymatically amplified DNA for the detection of pointmutations. Nucleic Acids Res 17:3606
Rohlfs EM, Learning WG, Friedman KJ et al (1997) Direst detection of mutations in the breast and ovarian cancer susceptibility gene BRCA1 by PCR-mediated site-directed mutagenesis. Clin Chem 43:24–29
Gayther SA, Harrington P, Russell P et al (1996) The UKCCCR Familial Ovarian Cancer Study Group. Rapid detection of regionally clustered germline BRCA1 mutations by multiplex heteroduplexanalysis. Am J Hum Genet 58:451–456
Friedman LS, Gayther SA, Kurosaki T et al (1997) Mutation analysis of BRCA1 and BRCA2 in a male breast cancer population. Am J Hum Genet 60:313–319
Ozcelik H, Antebi Y, Andrulis IL et al (1996) Heteroduplex and protein truncation analysis of the BRCA1 185delAG mutation. Hum Genet 98:310–312
Wagle N, Berger MF, Davis MJ et al (2012) High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov 2:82–93
Harismendy O, Schwab RB, Bao L et al (2011) Detection of low prevalence somatic mutations in solid tumors with ultra-deep targeted sequencing. Genome Biol 12:R124
Hadd AG, Houghton J, Choudhary A et al (2013) Targeted, high-depth, next-generation sequencing of cancer genes in formalin-fixed, paraffin-embedded and fine-needle aspiration tumor specimens. J Mol Diagn 15:234–247
Singh RR, Patel KP, Routbort MJ et al (2013) Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn 15:607–622
Loman NJ, Misra RV, Dallman TJ et al (2012) Performance comparison of benchtop high-throughput sequencing platforms.Nat. Biotech 30:434–439
Quail M, Smith M, Coupland P et al (2012) A tale of three next generation sequencing platforms: comparison of Ion torrent, pacific biosciences and illumina MiSeq sequencers. BMC Genom 13:341
Pellegrini L, Yu DS, Lo T et al (2002) Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420:287–293
Xia B, Sheng Q, Nakanishi K et al (2006) Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22:719–729
McCall CM, Stacy M et al (2014) False positives in multiplex PCR-based next-generation sequencing have unique signatures. J Mol Diagn 16(5):541–549
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Author declares no conflict of interest with any individual and/or financial organization.
Rights and permissions
About this article
Cite this article
Suryavanshi, M., Kumar, D., Panigrahi, M.K. et al. Detection of false positive mutations in BRCA gene by next generation sequencing. Familial Cancer 16, 311–317 (2017). https://doi.org/10.1007/s10689-016-9955-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-016-9955-8