Abstract
Resistance to lodging, an important problem in rice production, has three types: low plant height, strong culm, and high strength of the lower part of the plant. The determinants of strength of the lower part remains unclear, compared with plant height and culm strength. This study identified a new genetic factor involved in the strength of the lower part, as assessed by pushing resistance, using chromosomal segment substitution lines (CSSLs) to clarify the determinants of strength of the lower part by functional analysis of the CSSL and the introgression line (IL) harboring the identified quantitative trait locus (QTL). QTL analysis identified the QTL for increasing pushing resistance on chromosome 4, PRL4, which was not related to days to heading. The CSSL with PRL4 showed increased pushing resistance and physical strength of the basal culm, but decreased filled grain ratio and grain weight. The IL with PRL4, developed by backcrossing this CSSL, improved pushing resistance and the strain of culm until breaking under compression, and did not decrease yield traits. These lines with PRL4 increased the accumulation of non-structural carbohydrate (NSC) in the basal culm at the fully ripe stage. Thus, the genetic control of NSC accumulation in culms by PRL4 may improve the strength of the lower part by enhancing culm toughness with strength and ductility.



Similar content being viewed by others
Availability of data and materials
The datasets during and/or analysed during the current study available from the corresponding author on reasonable request.
Abbreviations
- CSSL:
-
Chromosomal segment substitution line
- IL:
-
Introgression line
- NSC:
-
Non-structural carbohydrate
- PRL:
-
Pushing resistance of the lower part
- PRL/TN:
-
Pushing resistance of the lower part per tiller
- QTL:
-
Quantitative trait locus
References
Bergmeyer HU, Bernt E (1974) Methods for determination of metabolites: carbohydrate metabolism: sucrose. In: Bergmeyer HU (ed) Methods of enzymatic analysis 3. Academic Press, New York, pp 1176–1179
Fan C, Li Y, Hu Z, Hu H, Wang G, Li A, Wang Y, Tu Y, Xia T, Peng L, Feng S (2018) Ectopic expression of a novel OsExtensin-like gene consistently enhances plant lodging resistance by regulating cell elongation and cell wall thickening in rice. Plant Biotechnol J 16:254–263
Guo Z, Liu X, Zhang B, Yuan X, Xing Y, Liu H, Luo L, Chen G, Xiong L (2021) Genetic analyses of lodging resistance and yield provide insights into post-Green Revolution breeding in rice. Plant Biotechnol J 19:814–829
Hirano K, Kotake T, Kamihara K, Tsuna K, Aohara T, Kaneko Y, Takatsuji H, Tsumuraya Y, Kawasaki S (2010) Rice BRITTLE CULM 3 (BC3) encodes a classical dynamin OsDRP2B essential for proper secondary cell wall synthesis. Planta 232:95–108
Hirano K, Okuno A, Hobo T, Ordonio R, Shinozaki Y, Asano K, Kitano H, Matsuoka M (2014) Utilization of stiff culm trait of rice smos1 mutant for increased lodging resistance. PLoS ONE 9:e96009
Hirano K, Ordonio RL, Matsuoka M (2017) Engineering the lodging resistance mechanism of post-Green Revolution rice to meet future demands. Proc Jpn Acad Ser B 93:220–233
International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436:793–800
Ishimaru K, Yano M, Aoki N, Ono K, Hirose T, Lin SY, Monna L, Sasaki T, Ohsugi R (2001) Toward the mapping of physiological and agronomic characters on a rice function map: QTL analysis and comparison between QTLs and expressed sequence tags. Theor Appl Genet 102:793–800
Ishimaru K, Togawa E, Ookawa T, Kashiwagi T, Madoka Y, Hirotsu N (2008) New target for rice lodging resistance and its effect in a typhoon. Planta 227:601–609
Kashiwagi T (2014) Identification of quantitative trait loci for resistance to bending-type lodging in rice (Oryza sativa L.). Euphytica 198:353–367
Kashiwagi T, Ishimaru K (2004) Identification and functional analysis of a locus for improvement of lodging resistance in rice. Plant Physiol 134:676–683
Kashiwagi T, Madoka Y, Hirotsu N, Ishimaru K (2006) Locus prl5 improves lodging resistance of rice by delaying senescence and increaseing carbohydrate reaccumulation. Plant Physiol Biochem 44:152–157
Kashiwagi T, Togawa E, Hirotsu N, Ishimaru K (2008) Improvement of lodging resistance with QTLs for stem diameter in rice (Oryza sativa L.). Theor Appl Genet 117:749–757
Kashiwagi T, Hirotsu N, Ujiie K, Ishimaru K (2010) Lodging resistance locus prl5 improves physical strength of the lower plant part under different conditions of fertilization in rice (Oryza sativa L.). Field Crops Res 115:107–115
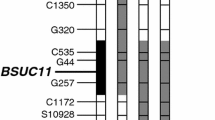
Kashiwagi T, Munakata J, Ishimaru K (2016) Functional analysis of the lodging resistance QTL BSUC11 on morphological and chemical characteristics in upper culms of rice. Euphytica 210:233–243
Kono M (1995) Physiological aspects of lodging. In: Matsuo T, Kumazawa K, Ishii R, Ishihara K, Hirata H (eds) Science of the rice plant, vol. 2: physiology. Food and Agriculture Policy Research Center, Tokyo, pp 971–982
Li X, Yang Y, Yao J, Chen G, Li X, Zhan Q, Wu C (2009) FLEXIBLE CULM1 encording a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol Biol 69:685–697
Madoka Y, Kashiwagi T, Hirotsu N, Ishimaru K (2008) Indian rice “Kasalath” contains genes that improve traits of Japanese premium rice “Koshihikari.” Theor Appl Genet 116:603–612
Merugumala GR, Satyanarayana PV, Narne C, Ravikumar BNVSR, Ramana Rao PV, Pavani L, Deepika V (2019) Molecular breeding of “Swarna”, a mega rice variety for lodging resistance. Mol Breed 39:55
Monna L, Kitazawa N, Yoshino R, Suzuki J, Masuda H, Maehara Y, Tanji M, Sato M, Nasu S, Minobe Y (2002) Positional cloning of rice semidwarfing gene, sd-1: rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res 9:11–17
Nomura T, Arakawa N, Yamamoto T, Ueda T, Adachi S, Yonemaru J-i et al (2019) Next generation long-culm rice with superior lodging resistance and high grain yield, Monster Rice 1. PLoS ONE 14:e0221424
Ookawa T, Ishihara K (1992) Varietal difference of physical characteristics of the culm related to lodging resistance in paddy rice. Jpn J Crop Sci 61:419–425
Ookawa T, Hobo T, Yano M, Murata K, Ando T, Miura H, Asano K, Ochiai Y, Ikeda M, Nishitani R, Ebitani T, Ozaki H, Angeles ER, Hirasawa T, Matsuoka M (2010a) New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat Commun 1:132
Ookawa T, Yasuda K, Kato H, Sakai M, Seto M, Sunaga K, Motobayashi T, Tojo S, Hirasawa T (2010b) Biomass production and lodging resistance in ‘Leaf Star’, a new long-culm rice forage cultivar. Plant Prod Sci 13:58–66
Peng S, Cassman K, Virmani SS, Sheehy J, Khush GS (1999) Yield potential trends of tropical rice since the release of IR8 and the challenge of increasing rice yield potential. Crop Sci 39:1552–1559
Sowadan O, Li D, Zhang Y, Zhu S, Hu X, Bhanbhro LB, Edzesi WM, Dang X, Hong D (2018) Mining of favorable alleles for lodging resistance traits in rice (Oryza sativa) through association mapping. Planta 248:155–169
Suzuki S, Suzuki Y, Yamamoto N, Hattori T, Sakamoto M, Umezawa T (2009) High-throughput determination of thioglycolic acid lignin from rice. Plant Biotechnol 26:337–340
Takai T, Nonoue Y, Yamamoto S, Yamanouchi U, Matsubara K, Liang ZW, Lin H, Ono N, Uga Y, Yano M (2007) Development of chromosome segment substitution lines derived from backcross between indica donor rice cultivar ‘Nona Bokra’ and japonica recipient cultivar ‘Koshihikari.’ Breed Sci 57:257–261
Terashima K, Ogata T, Akita S (1994) Eco-physiological characteristics related with lodging tolerance of rice in direct sowing cultivation. II. Root growth characteristics of tolerant cultivars to root lodging. Jpn J Crop Sci 63:34–41
Ujiie K, Kashiwagi T, Ishimaru K (2012) Identification and functional analysis of alleles for productivity in two sets of chromosome segment substitution lines of rice. Euphytica 187:325–337
Wise LE, Murphy M, D’Addieco AA (1946) Chlorite holocellulose, its fractionation and bearing on summative wood analysis and studies on the hemicelluloses. Paper Trade J 122:35–43
Yamauchi K, Yamamoto T, Segami S, Horikawa M, Chaya G, Kitano H, Iwasaki Y, Miura K (2020) gw2 mutation increases grain width and culm thickness in rice (Oryza sativa L.). Breed Sci 70:456–461
Yano K, Ookawa T, Aya K, Ochiai Y, Hirasawa T, Ebitani T, Takarada T, Yano M, Yamamoto T, Fukuoka S, Wu J, Ando T, Ordonio RL, Hirano K, Matsuoka M (2015) Isolation of a novel lodging resistance QTL gene involved in Strigolactone signaling and its pyramiding with a QTL gene involved in another mechanism. Mol Plant 8:303–314
Acknowledgements
Not applicable.
Funding
This work was supported by JSPS KAKENHI Grant Number JP20K05993.
Author information
Authors and Affiliations
Contributions
The concept, experimental design, analysis, writing, and revision of the manuscript were conducted by TK.
Corresponding author
Ethics declarations
Conflict of interest
The author declares that there are no conflict of interest.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kashiwagi, T. Novel QTL for lodging resistance, PRL4, improves physical properties with high non-structural carbohydrate accumulation of basal culms in rice (Oryza sativa L.). Euphytica 218, 83 (2022). https://doi.org/10.1007/s10681-022-03036-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-022-03036-6