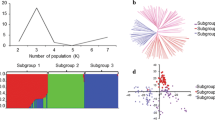
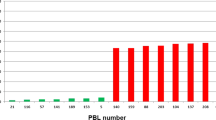
Abstract
Genetic resistance to common bunt is a cost-effective, environmentally friendly, and sustainable approach to controlling the disease. To date, 16 race specific common bunt resistance genes (Bt1-Bt15 and Btp) have been reported in wheat. However, a limited number have been mapped and few markers have been identified, which limits the usage of molecular markers in a marker-assisted breeding program. A total of 125 synthetic hexaploid wheats (SHWs) were evaluated for reactions to a mixture of common bunt races under field conditions in Turkey in 2016 and 2017. The objectives of this study were to identify common bunt resistant genotypes, identify genomic regions conferring resistance to common bunt using 35,798 genotyping-by-sequencing derived single nucleotide polymorphisms (SNPs), and investigate the significant SNPs present within genes using the functional annotations of the underlying genes. We found 29 resistant SHWs that can be used in wheat breeding. The genome-wide association study identified 15 SNPs associated with common bunt resistance and a haplotype block comprising three SNPs in perfect linkage disequilibrium. Five of them were novel and were located on chromosomes 2A, 3D, and 4A. Furthermore, seven of the 15 SNPs were found within genes and had annotations suggesting potential role in disease resistance. This study identified several favorable alleles that decreased common bunt incidence up to 26% in SHWs. These resistant SHWs and candidate genomic regions controlling common bunt resistance will be useful for wheat genetic improvement and could assist in further understanding of the genetic architecture of common bunt resistance.





Similar content being viewed by others
Abbreviations
- SHW:
-
Synthetic hexaploid wheat
- GBS:
-
Genotyping-by-sequencing
- MTA:
-
Marker-trait association
- BLUP:
-
Best linear unbiased predictor
- GWAS:
-
Genome-wide association study
- MAF:
-
Minor allele frequency
- Q–Q plot:
-
Quantile–quantile plot
- SNP:
-
Single nucleotide polymorphism
- IWGSC:
-
International Wheat and Genome Sequencing Consortium
References
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. https://doi.org/10.1093/bioinformatics/bth457
Bhatta M, Regassa T (2018) Seed treatment affected yield and economic returns of Nebraska winter wheat genotypes. J Agri Sci 9:5–10
Bhatta M, Morgounov A, Belamkar V et al (2018a) Unlocking the novel genetic diversity and population structure of synthetic hexaploid wheat. BMC Genom 19:951. https://doi.org/10.1186/s12864-018-4969-2
Bhatta M, Morgounov A, Belamkar V, Baenziger PS (2018b) Genome-wide association study for grain yield and yield-related traits in drought-stressed synthetic hexaploid wheat germplasm. Int J Mol Sci 19:3011. https://doi.org/10.3390/ijms19103011
Bozkurt O, Unver T, Akkaya MS (2007) Genes associated with resistance to wheat yellow rust disease identified by differential display analysis. Physiol Mol Plant Pathol 71:251–259. https://doi.org/10.1016/j.pmpp.2008.03.002
Bradbury PJ, Zhang Z, Kroon DE et al (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Chen B, Wang J, Zhang G, Liu J, Manan S, Hu H, Zhao J (2016) Two types of soybean diacylglycerol acyltransferases are differentially involved in triacylglycerol biosynthesis and response to environmental stresses and hormones. Sci Rep 6:28541. https://doi.org/10.1038/srep28541
Cheung M-Y, Zeng N-Y, Tong S-W et al (2007) Expression of a RING-HC protein from rice improves resistance to Pseudomonas syringae pv. tomato DC3000 in transgenic Arabidopsis thaliana. J Exp Bot 58:4147–4159. https://doi.org/10.1093/jxb/erm272
Ciucă M (2011) A preliminary report on the identification of SSR markers for bunt (Tilletia sp.) resistance in wheat. Czech J Genet Plant Breed 47:S142–S145. https://doi.org/10.17221/3269-CJGPB
Dumalasová V, Simmonds J, Bartoš P, Snape J (2012) Location of genes for common bunt resistance in the European winter wheat cv. Trintella. Euphytica 186:257–264. https://doi.org/10.1007/s10681-012-0671-7
Fofana B, Humphreys DG, Cloutier S et al (2008) Mapping quantitative trait loci controlling common bunt resistance in a doubled haploid population derived from the spring wheat cross RL4452 × AC Domain. Mol Breed 21:317–325. https://doi.org/10.1007/s11032-007-9131-9
Galaev AV, Babayants LT, Sivolap YM (2012) DNA-markers for resistance to common bunt transferred from Aegilops cylindrica Host. to hexaploid wheat. Czech J Genet Plant Breed 42:62–65. https://doi.org/10.17221/6234-CJGPB
Glaubitz JC, Casstevens TM, Lu F et al (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS ONE 9:e90346. https://doi.org/10.1371/journal.pone.0090346
Goates BJ (2012) Identification of new pathogenic races of common bunt and dwarf bunt fungi, and evaluation of known races using an expanded set of differential wheat lines. Plant Dis 96:361–369. https://doi.org/10.1094/PDIS-04-11-0339
Goates BJ, Bockelman HE (2012) Identification of new sources of high levels of resistance to dwarf bunt and common bunt among winter wheat landraces in the USDA-ARS national small grains collection. Crop Sci 52:2595. https://doi.org/10.2135/cropsci2012.01.0060
He C, Hughes GR (2003) Inheritance of resistance to common bunt in spelt and common wheat. Can J Plant Sci 83:47–56. https://doi.org/10.4141/P01-167
Hong JK, Choi HW, Hwang IS, Hwang BK (2007) Role of a novel pathogen-induced pepper C3–H–C4 type RING-finger protein gene, CaRFP1, in disease susceptibility and osmotic stress tolerance. Plant Mol Biol 63:571–588. https://doi.org/10.1007/s11103-006-9110-2
Im S, Lee H-N, Jung HS et al (2017) Transcriptome-based identification of the desiccation response genes in marine red algae Pyropia tenera (Rhodophyta) and enhancement of abiotic stress tolerance by PtDRG2 in chlamydomonas. Mar Biotechnol 19:232–245. https://doi.org/10.1007/s10126-017-9744-x
International Wheat Genome Sequencing Consortium (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361(6403):eaar7191. https://doi.org/10.1126/science.aar7191
Knox RE, Campbell HL, Depauw RM et al (2013) DNA markers for resistance to common bunt in ‘McKenzie’ wheat. Can J Plant Pathol 35:328–337. https://doi.org/10.1080/07060661.2013.763292
Li W-T, Chen W-L, Yang C et al (2014) Identification and network construction of zinc finger protein (ZFP) genes involved in the rice-‘Magnaporthe oryzae’ interaction. Plant Omics 7:540
Liu X, Huang M, Fan B et al (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12:e1005767. https://doi.org/10.1371/journal.pgen.1005767
McIntosh RA (1998) Catalogue of gene symbols for wheat. In: Proceedings of the 9th international wheat genetics symposium. University of Saskatchewan, Saskatoon, Canada 1998, vol 5, pp 119–120
Menzies JG, Knox RE, Popovic Z, Procunier JD (2006) Common bunt resistance gene Bt10 located on wheat chromosome 6D. Can J Plant Sci 86:1409–1412. https://doi.org/10.4141/P06-106
Moore AL, Shiba T, Young L et al (2013) Unraveling the heater: new insights into the structure of the alternative oxidase. Annu Rev Plant Biol 64:637–663. https://doi.org/10.1146/annurev-arplant-042811-105432
Morgounov A, Abugalieva A, Akan K et al (2018) High-yielding winter synthetic hexaploid wheats resistant to multiple diseases and pests. Plant Genet Resour 16:273–278. https://doi.org/10.1017/S147926211700017X
Niu J, Jia H, Yin J et al (2010) Development of an STS marker linked to powdery mildew resistance genes PmLK906 and Pm4a by gene chip hybridization. Agric Sci China 9:331–336. https://doi.org/10.1016/S1671-2927(09)60101-2
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
SAS Institute (2018) SAS 9.4 product documentation. SAS Institute, Cary, NC. http://support.sas.com/documentation/94/index. html. Accessed 10 May 2018
Schaller CW, Holton CS, Kendrick EL (1960) Inheritance of the second factor for resistance to bunt, Tilletia caries and T. foetida, in the wheat variety Martin. Agron J 52:280–285
Scmidt JW, Morris R, Johnson VA (1969) Monosomic analysis for bunt resistance in derivatives of Turkey and oro wheats 1. Crop Sci 9:286–288. https://doi.org/10.2135/cropsci1969.0011183X000900030009x
Sears ER, Schaller CW, Briggs FN (1960) Identification of the chromosome carrying the Martin gene for resistance of wheat to bunt. Can J Genet Cytol 2:262–267. https://doi.org/10.1139/g60-026
Singh A, Knox RE, DePauw RM et al (2016) Genetic mapping of common bunt resistance and plant height QTL in wheat. Theor Appl Genet 129:243–256. https://doi.org/10.1007/s00122-015-2624-8
Steffan PM, Torp AM, Borgen A et al (2017) Mapping of common bunt resistance gene Bt9 in wheat. Theor Appl Genet 130:1031–1040. https://doi.org/10.1007/s00122-017-2868-6
Vanlerberghe G (2013) Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int J Mol Sci 14:6805–6847. https://doi.org/10.3390/ijms14046805
Veisz O, Szunics L, Szunics L (2000) Effect of common bunt on the frost resistance and winter hardiness of wheat (Triticum aestivum L.) lines containing Bt genes. Euphytica 114:159–164. https://doi.org/10.1023/A:1003940808041
Wang S, Knox RE, DePauw RM et al (2009) Markers to a common bunt resistance gene derived from ‘Blizzard’ wheat (Triticum aestivum L.) and mapped to chromosome arm 1BS. Theor Appl Genet 119:541–553. https://doi.org/10.1007/s00122-009-1063-9
Zhu J, Lee B-H, Dellinger M et al (2010) A cellulose synthase-like protein is required for osmotic stress tolerance in Arabidopsis: SOS6 is important for osmotic stress tolerance in plants. Plant J 63:128–140. https://doi.org/10.1111/j.1365-313X.2010.04227.x
Zou J, Semagn K, Chen H et al (2017) Mapping of QTLs associated with resistance to common bunt, tan spot, leaf rust, and stripe rust in a spring wheat population. Mol Breed 37:144. https://doi.org/10.1007/s11032-017-0746-1
Acknowledgements
This work was funded by the Monsanto Beachell-Borlaug International Scholarship Program. This project is a collaborative effort between the University of Nebraska-Lincoln, Lincoln, USA, the International Maize and Wheat Improvement Center (CIMMYT) in Turkey (supported by CRP WHEAT; Ministry of Food, Agriculture and Livestock of Turkey; Bill and Melinda Gates Foundation, and UK Department for International Development, grant OPP1133199), and Omsk State Agrarian University (supported by the Russian Science Foundation project No. 16-16-10005). We would like to thank CIMMYT-Turkey for providing seeds of synthetic hexaploid wheat and Dr. Jesse Poland for providing the GBS data. We would like to acknowledge CIMMYT-Turkey staffs including Dr. Amer Dabadat, Dr. Gul Erginbas-Orakci, Adem Urglu, Ibrahim Ozturk, and the Transitional Zone Agricultural Research Institute, Eskisehir, Turkey for providing experimental station and assisted on managing common bunt experiments. Partial funding for P.S. Baenziger is from Hatch project NEB-22-328, AFRI/2011-68002-30029, the USDA National Institute of Food and Agriculture as part of the International Wheat Yield Partnership award number 2017-67007-25939, the CERES Trust Organic Research Initiative, and USDA under Agreement No. 59-0790-4-092 which is a cooperative project with the U.S. Wheat and Barley Scab Initiative. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the view of the USDA. Cooperative investigations of the Nebraska Agric. Res. Div., Univ. of Nebraska, and USDA-ARS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bhatta, M., Morgounov, A., Belamkar, V. et al. Genome-wide association study reveals favorable alleles associated with common bunt resistance in synthetic hexaploid wheat. Euphytica 214, 200 (2018). https://doi.org/10.1007/s10681-018-2282-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-018-2282-4