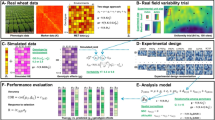
Abstract
Field variation is one of the important factors that can have a significant impact on genetic data analysis. Ineffective control of field variation may result in an inflated residual variance and/or biased estimation of genetic variations and/or effects. In this study, we addressed this problem by merging genetic models with the information from a rectangular cotton field layout (referred to row and column directions). Data from a genetic mapping study in Upland cotton (Gossypium hirsutum L.) was used to validate the proposed methodology. This study included model evaluation based on simulations and actual data analysis on four agronomic traits (seed yield, lint yield, lint percentage, and boll weight) in cotton. Results based on simulations suggested that when there were no row and column effects, the conventional and the extended genetic models yielded similar results. However, when either field row and/or column effects were significant, the conventional genetic model yielded biased estimates for residual variance component with larger mean square error whereas the extended genetic models yielded more unbiased estimates. Actual data analysis revealed that lint yield and seed yield were significantly influenced by the systematic variation present in the field. With the extended model, the residual variance associated with these traits was reduced approximately 65 % compared to the conventional block model. Accordingly, the averaged heritability estimate increased by about 18 % for these traits. Thus, the results suggested that genetic data analysis can be improved when field variation is considered.



Similar content being viewed by others
References
Besag J, Kempton R (1986) Statistical analysis of field experiments using neighbouring plots. Biometrics 42:231–251
Cullis B, Gogel B, Verbyla A, Thompson R (1998) Spatial analysis of multi-environment early generation variety trials. Biometrics 54:1–18
Federer WT (1998) The recovery of interblock, intergradient, and intervariety information in incomplete block and lattice rectangle designs experiments. Biometrics 54:471–481
Federer WT (2003) Exploratory model selection for spatially designed experiments- some examples. J Data Sci 1:231–248
Federer WT, Nair RC, Raghavarao D (1975) Some augmented row-column designs. Biometrics 31:361–373
Fehr W (1987) Principles of cultivar development: theory and technique, vol 1. Macmillan Publishing Co., New York
Gleeson AC, Cullis BR (1987) Residual maximum likelihood (REML) estimation of neighbour model for field experiments. Biometrics 43:277–287
Hartley HO, Rao JNK (1967) Maximum-likelihood estimation for the mixed analysis of variance model. Biometrika 54:93–108
Jenkins J, McCarty J Jr, Wu J, Gutierrez O (2009) Genetic variance components and genetic effects among eleven diverse upland cotton lines and their F2 hybrids. Euphytica 167:397–408
Kempton RA, Seraphin JC, Sword AM (1994) Statistical analysis of two-dimensional variation in variety yield trials. J Agric Sci 122:335–342
Nguyen N-K, Williams ER (1993) An algorithm for constructing optimal resolvable row-column designs. Aust J Stat 35:363–370
Patterson HD, Hunter EA (1983) The efficiency of incomplete block designs in National List and Recommended List cereal variety trials. J Agric Sci 101:427–433
Qiao CG, Basford KE, DeLacy IH, Cooper M (2000) Evaluation of experimental designs and spatial analyses in wheat breeding trials. Theor Appl Genet 100:9–16
Qureshi SN, Saha S, Kantety RV, Jenkins JN (2004) EST-SSR: a new class of genetic markers in cotton. J Cotton Sci 8:112–123
Rao CR (1971) Estimation of variance and covariance components—MINQUE theory. J Multivar Anal 1:257–275
Robinson DL, Kershaw CD (1988) An investiation of two-dimensional yield variability in breeders’ small plot barley trials. J Agr Sci 111:419–426
Searle SR, Casella G, McCulloch CE (1992) Variance components. Wiley, New York
Shappley ZW, Jenkins JN, Watson CE, Kahler AL, Meredith WR (1996) Establishment of molecular markers and linkage groups in two F-2 populations of Upland cotton. Theor Appl Genet 92:915–919
Williams ER (1986) A neighbour model for field experiments. Biometrika 73:279–287
Wu J (2014) minque: an R package fir linear mixed model analyses. 1.1 edn, http://cran.r-project.org/
Wu J, Xu H, Zhu J (1998) An approach to eliminating systematic errors in genetic analysis. In: Chen LS, Ruan SG, Zhu J (eds) International conference on mathematical biology. World Scientific Publishings Co., Singapore, pp 265–270
Wu J, Jenkins JN, McCarty JC, Zhu J (2004) Genetic association of yield with its component traits in a recombinant inbred line population of cotton. Euphytica 140:171–179
Wu J, Jenkins JN, McCarty JC, Saha S, Stelly DM (2006a) An additive-dominance model to determine chromosomal effects in chromosome substitution lines and other gemplasms. Theor Appl Genet 112:391–399
Wu J, Jenkins JN, McCarty JC, Wu D (2006b) Variance component estimation using the additive, dominance, and additive × additive model when genotypes vary across environments. Crop Sci 46:174–179
Wu J, Bondalapati K, Glover K, Berzonsky W, Jenkins JN, McCarty JC (2013) Genetic analysis without replications: model evaluation and application in spring wheat. Euphytica 190:447–458
Zhu J (1989) Estimation of genetic variance components in the general mixed model. North Carolina State, Raleigh
Zhu J (1993) Methods of predicting genotype value and heterosis for offspring of hybrids. J Biomath 8:32–40
Zimmerman DL, Harville DA (1991) A random field approach to the analysis of field-plot experiments and other spatial experiments. Biometrics 47:223–239
Acknowledgments
This research was partially supported by funding USDA-NIFA Hatch project 1005459 and the South Dakota Agricultural Experiment Station.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bondalapati, K.D., Jenkins, J.N., McCarty, J.C. et al. Field experimental design comparisons to detect field effects associated with agronomic traits in upland cotton. Euphytica 206, 747–757 (2015). https://doi.org/10.1007/s10681-015-1512-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-015-1512-2