Abstract
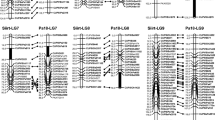
Populus adenopoda Maxim. and P. alba L. [section Populus (aspen), genus Populus] are two tree species of ecological and economic value. To date, no high-density genetic maps are available for these two species. In this study, 1100 interspecific hybrids were obtained by controlled crossing and embryo culture. Simple sequence repeat (SSR) and sequence-related amplified polymorphisms (SRAP) were used to genotype 189 F1 individuals. The genetic linkage map of P. adenopoda × P. alba generated from this study includes 212 markers (192 SSRs and 50 SRAPs) and consists of 26 linkage groups spanning 2178.5 cM, with an average distance of 11.7 cM between markers. This is the first SSR- and SRAP-containing genetic linkage map for aspen. The SSRs on the map will serve both as bridges for comparison with the poplar maps published to date and as a direct link to the Populus genomic sequence. Future studies focusing on the data presented here should enhance the density and precision of the map for identifying and localizing quantitative trait loci and promote genomic research on the genus.


Similar content being viewed by others
References
Bradshaw HD, Villar M, Watson BD, Otto KG, Stewart S, Stettler RF (1994) Molecular genetics of growth and development in Populus. III. A genetic linkage map of a hybrid poplar composed of RFLP, STS, and RAPD markers. Theor Appl Genet 89:167–178
Bradshaw HD, Ceulemans R, Davis J, Stettler R (2000) Emerging model systems in plant biology: poplar (Populus) as a model forest tree. J Plant Growth Regul 19:306–313. doi:10.1007/s003440000030
Brunner AM, Rottmann WH, Sheppard LA, Krutovski K, DiFazio SP, Leonardi S, Strauss SH (2000) Structure and expression of duplicate AGAMOUS orthologous in poplar. Plant Mol Biol 44:619–634
Budak H, Shearman RC, Parmaksiz I, Gaussoin RE, Riordan TP, Dweikat I (2004) Molecular characterization of Buffalograss germplasm using sequence_related amplified polymorphism markers. Theor Appl Genet 108:328–334. doi:10.1007/s00122-003-1428-4
Cervera MT, Storme V, Ivens B, Gusmao J, Liu BH, Hostyn V, Van Slycken J, Van Montagu M, Boerjan W (2001) Dense genetic linkage maps of three Populus species (Populus deltoides, P. nigra and P. trichocarpa) based on AFLP and microsatellite markers. Genetics 158:787–809
Cervera MT, Sewell MM, Faivre-Rampant P, Storme V, Boerjan W (2004) Genome mapping in Populus. In: Kumar S, Fladung M (eds) Molecular genetics and breeding of forest trees. Food Product Press, New York, pp 387–410
Cervera MT, Storme V, Soto A, Ivens B, Van Montagu M, Rajora OP, Boerjan W (2005) Intraspecific and interspecific genetic and phylogenetic relationships in the genus Populus based on AFLP markers. Theor Appl Genet 111:1440–1456. doi:10.1007/s00122-005-0076-2
Cole CT (2005) Allelic and population variation of microsatellite loci in aspen (Populus tremuloides). New Phytol 167:155–164. doi:10.1111/j.1469-8137.2005.01423.x
Collard BCY, Pang ECK, Jahufer MZZ, Brouwer JB (2005) An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142:169–196. doi:10.1007/s10681-005-1681-5
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Eckenwalder JE (1996) Systematic and evolution of Populus. In: Stettler RF, Bradshaw HD Jr, Heilman PE, Hinckley TM (eds) Biology of Populus and its implications for management and conservation. NRC Research Press, Ottawa, pp 7–32
Ferriol M, Pico B, Nuez F (2003) Genetic diversity of a germplasm collection of Cucurbita pepo using SARP and AFLP markers. Theor Appl Genet 107:271–282. doi:10.1007/s00122-003-1242-z
Ferriol M, Pico B, Fernandez de Cordova P, Nuez F (2004) Molecular diversity of a germplasm collection of squash (Cucurbita moschata) determined by SRAP and AFLP markers. Crop Sci 44:653–664
Fu XP, Ning GG, Gao LP, Bao MZ (2008) Genetic diversity of Dianthus accessions as assessed using two molecular marker systems (SRAPs and ISSRs) and morphological traits. Sci Hortic 117:263–270. doi:10.1016/j.scienta.2008.04.001
Gaudet M (2006) Molecular approach to dissect adaptive traits in native European Populus nigra L. construction of a genetic linkage map based on AFLP, SSR, and SNP markers. http://hdl.handle.net/2067/99
Gaudet M, Jorge V, Paolucci I, Beritognolo I, Scarascia Mugnozza G, Sabatti M (2007) Genetic linkage maps of Populus nigra L. including AFLPs, SSRs, SNPs, and sex trait. Tree Genet Genomes 4:25–36. doi:10.1007/s11295-007-0085-1
Gerber S, Rodolphe F (1994) An estimation of the genome length of maritime pine (Pinus pinaster Ait). Theor Appl Genet 88:289–292. doi:10.1007/BF00223634
Hamzeh M, Dayanandan S (2004) Phylogeny of Populus (Salicaceae) based on nucleotide sequences of chloroplast trnT-trnF region and nuclear rDNA. Am J Bot 91:1398–1408
Hanley SJ, Mallott MD, Karp A (2006) Alignment of a Salix linkage map to the Populus genomic sequence reveals microsynteny between willow and poplar genomes. Tree Genet Genomes 3:35–48. doi:10.1007/s11295-006-0049-x
Hao Q, Liu ZA, Shu QY, Zhang R, DeRick J, Wang LS (2008) Studies on Paeonia cultivars and hybrids identification based on SRAP analysis. Hereditas 145:38–47. doi:10.1111/j.0018-0661.2008.2013.x
Hulbert SH, Ilott TW, Legg EJ, Lincoln SE, Lander ES, Michelmore RW (1988) Genetic analysis of the fungus, Bremia lactucae, using restriction fragment length polymorphisms. Genetics 120:947–958
Jansson S, Douglas CJ (2007) Populus: a model system for plant biology. Annu Rev Plant Biol 58:435–458. doi:10.1146/annurev.arplant.58.032806.103956
Kuang H, Richardson T, Carson S, Wilcox P, Bongarten B (1999) Genetic analysis of inbreeding depression in plus tree 850.55 of Pinus radiata D. Don. I. Genetic map with distorted markers. Theor Appl Genet 98:697–703. doi:10.1007/s001220051123
Lange K, Boehnke M (1982) How many polymorphic genes will it take to span the human genome? Am J Hum Genet 24:842–845
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SARP) a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103:455–461. doi:10.1007/s001220100570
Li G, Gao M, Yang B, Quiros CF (2003) Gene for gene alignment between the Brassica and Arabidopsis genomes by direct transcriptome mapping. Theor Appl Genet 107:168–180. doi:10.1007/s00122-003-1236-x
Lin ZX, Zhang XL, Nie YC (2004) Evaluation on of application of a new molecular marker SRAP analysis of F2 segregation population and genetic diversity in cotton (in Chinese). Acta Gent Sin 31:622–626
Liu E (2008) Construction of a frame genetic map for Salix suchowensis × S. erioclada L. Master thesis, Nanjing Forestry University, China
Oetting WS, Lee HK, Flanders DJ, Wiesner GL, Sellers TA, King RA (1995) Linkage analysis with multiplexed short tandem repeat polymorphisms using infrared fluorescence and M13 tailed primers. Genomics 30:450–458. doi:10.1006/geno.1995.1264
Pakull B, Groppe K, Meyer M, Markussen T, Fladung M (2009) Genetic linkage mapping in aspen (Populus tremula L. and Populus tremuloides Michx.). Tree Genet Genomes 5:505–515. doi:10.1007/s11295-009-0204-2
Pan QH, Hu ZD, Tanisaka T, Wang L (2003) Fine mapping of the blast resistance gene Pil5, linked to Pii on rice chromosome 9. Acta Bot Sin 45:871–877
Remington DL, Whetten RW, Liu BH, O’Malley DM (1999) Construction of an AFLP genetic map with nearly complete genome coverage in Pinus taeda. Theor Appl Genet 98:1279–1292. doi:10.1007/s001220051194
Riaz A, Potter D, Stephen SM (2004) Genotyping of peach and nectarine cultivars with SSR and SRAP molecular markers. J Am Soc Hortic Sci 129:204–211
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234. doi:10.1038/72708
Smulders MJM, Van Der Schoot J, Arens P, Vosman B (2001) Trinucleotide repeat microsatellite markers for black poplar (Populus nigra L.). Mol Ecol Notes 1:188–190. doi:10.1046/j.1471-8278.2001.00071.x
Stam P (1993) Construction of integrated genetic linkage maps by means by a new computer package: JOINMAP. Plant J 3:739–744
Sun Z, Wang Z, Tu J, Zhang J, Yu F, Peter BE, Li G (2007) An ultradense genetic recombination map for Brassica napus, consisting of 13551 SRAP markers. Theor Appl Genet 114:1305–1317. doi:10.1007/s00122-006-0483-z
Taylor G (2002) Populus: arabidopsis for forestry. Do we need a model tree? Ann Bot (Lond) 90:681–689. doi:10.1093/aob/mcf255
Tuskan GA, DiFazio SP, Teichmann T (2003) Poplar genomics is getting popular: the impact of the poplar genome project on tree research. Plant Biol 5:1–3
Tuskan GA, Gunter LE, Yang ZM, Yin TM, Sewell MM, Difazio SP (2004) Characterization of microsatellites revealed by genomic sequencing of Populus trichocarpa. Can J For Res 34:85–93. doi:10.1139/x03-283
Tuskan GA, Jansson S, Bohlmann J et al (2006) The genome of black cottonwood, Populus trichocarpa (Tort. & Gray). Science 313:1596–1604. doi:10.1126/science.1128691
Van der Schoot J, Pospíšková M, Vosman B, Smulders MJM (2000) Development and characterization of microsatellite markers in black poplar (Populus nigra L.). Theor Appl Genet 101:317–322. doi:10.1007/s001220051485
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. doi:10.1093/jhered/93.1.77
Woolbright SA, DiFazio SP, Yin T, Martinsen GD, Zhang X, Allan GJ, Whitham TG, Keim P (2008) A dense linkage map of hybrid cottonwood (Populus fremontii × P. angustifolia) contributes to long-term ecological research and comparison mapping in a model forest tree. Heredity 100:59–70. doi:10.1038/sj.hdy.6801063
Xu HY (1988) Poplars. Heilongjinag People Press, Haerbian
Yeboah MA, Xeuhao C, Feng CR, Alfandi M, Liang G, Gu M (2008) Mapping quantitative trait loci for waterlogging tolerance in cucumber using SRAP and ISSR markers. Biotechnology 7(2):157–167
Yin TM, Huang MR, Wang MX, Zhu LH, Zeng ZB, Wu RL (2001) Preliminary interspecific genetic maps of the Populus genome constructed from RAPD markers. Genome 44:602–609. doi:10.1139/gen-44-4-602
Yin TM, DiFazio SP, Gunter LE, Riemenschneider D, Tuskan GA (2004) Large-scale heterospecific segregation distortion in Populus revealed by a dense genetic map. Theor Appl Genet 109:451–463. doi:10.1007/s00122-004-1653-5
Zhang DQ, Zhang ZY, Yang K, Tian L (2002) Segregation of AFLP markers in a (Populus tomentosa × P. bolleana) × P. Tomen tosa Carr. BC1 family. For Stud China 4:21–26
Zhang B, Zhang L, Zhuge Q, Wang MX, Huang MR (2004) A rapid and simple method of total DNA extraction from tree (in Chinese). J Nanjing For Univ 28:13–16
Zhang B, Tong C, Yin T, Zhang X, Zhuge Q, Huang M, Wang M, Wu R (2009) Detection of quantitative trait loci influencing growth trajectories of adventitious roots in Populus using functional mapping. Tree Genet Genomes 5:539–552. doi:10.1007/s11295-009-0207-z
Acknowledgments
We thank the anonymous reviewers for their constructive comments on this manuscript. This work was supported by the National High Technology Research and Development Program of China (863 Program: 2006AA100109) and Key Laboratory of Forest Genetics & Biotechnology (Nanjing Forestry University), Ministry of Education of China (Grant No. FGB200802).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, Y., Sun, X., Tan, B. et al. A genetic linkage map of Populus adenopoda Maxim. × P. alba L. hybrid based on SSR and SRAP markers. Euphytica 173, 193–205 (2010). https://doi.org/10.1007/s10681-009-0085-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-009-0085-3