Abstract
Parastagonospora nodorum leaf and glume blotch (syn. Septoria nodorum blotch, SNB) is a severe disease in many wheat-growing areas worldwide. In a previous study, a mapping population, Liwilla × Begra, was used to detect several resistance quantitative trait loci (QTL) at the seedling stage. In this study the same mapping population was analysed at the adult plant stage under field and polytunnel conditions. After artificial inoculation the disease severity on leaves and glumes was scored as the areas under the disease progress curves for field tests and as the percentage of the leaf and glume area covered by necrosis for the polytunnel test. Three QTL associated with Septoria nodorum glume blotch resistance and two QTL associated with Septoria nodorum leaf blotch resistance were detected on chromosomes 1B, 3A, 4A and 7D. Each of the detected QTL explained only a small proportion of the total phenotypic variation, ranging from 9.1 to 20.0%. None of these QTL co-located with necrotrophic effector sensitivity loci or aligned with previously identified resistance loci at the seedling stage for the Liwilla × Begra population. SNB resistance QTL detected in our study did not overlap with QTL associated with morphological and developmental traits. Therefore they could be involved in the defence reaction and can be considered in wheat improvement for SNB resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Parastagonospora (syn. Septoria, syn. Ana Stagonospora, teleomorph Phaeosphaeria) nodorum (Berk.) Quaedvlieg, Verkley and Crous is the causal agent of Septoria nodorum blotch (SNB), an important disease of wheat worldwide (Solomon et al. 2006; Quaedvlieg et al. 2013). This necrotrophic fungus is able to infect both glumes and leaves, reducing grain yield and quality. Yield losses can reach 50% or more during severe epidemics (Bostwick et al. 1993; Shaner and Buechley 1995; Bhathal et al. 2003). Intensified wheat production, reduced tillage, shorter crop rotation and the use of susceptible cultivars are the most common factors contributing to the disease severity. The major primary inoculum are the wind-dispersed ascospores (sexual form) in late autumn and early spring (Keller et al. 1997; Bathgate and Loughman 2001). Asexually produced, rain-splashed pycnidiospores contribute to the development of a disease epidemic during the growing season, especially under warm, wet weather conditions (Arseniuk et al. 1998). Disease severity may be affected by plant height and maturity because the flag leaves and glumes, important in grain filling, are infected with pycnidiospores earlier on short and early maturing plants than on tall and later maturing plants (Eyal 1981; Scott et al. 1982). Sexual recombination of the fungus contributes to high genetic variation, which may allow the pathogen to adapt rapidly to a resistant cultivar or a fungicide (McDonald and Linde 2002; Blixt et al. 2009; Sommerhalder et al. 2010). The most effective, ecological and economical method of controlling SNB is the use of host resistance (Francki 2013).
Most studies suggest that inheritance of SNB resistance is complex and under polygenic control (Fried and Meister 1987; Bostwick et al. 1993; Du et al. 1999), but monogenic inheritance has also been observed in some wheat genotypes (reviewed by Xu et al. 2004; Friesen et al. 2008a). Usually, additive and dominant effects, and less frequently epistatic interactions, play a role in controlling seedling resistance (Mullaney et al. 1982; Ecker et al. 1989; Wilkinson et al. 1990). Adult plant resistance is similarly inherited but usually under the control of different genes for leaf and glume resistance (Rufty et al. 1981; Nelson and Gates 1982; Fried and Meister 1987; Bostwick et al. 1993; Wicki et al. 1999).
Molecular mapping and quantitative trait loci (QTL) analyses using bi-parental populations and genome-wide association studies (GWAS) greatly augment the understanding of the inheritance and genetic control of SNB resistance. A number of QTL have been identified on chromosomes such as 1A, 1B, 2B, 2D, 3A, 4A, 4B, 4D, 5A, 5B, 5D, 6A, 6D, 7A, 7B and 7D for the seedling resistance (Czembor et al. 2003; Arseniuk et al. 2004; Liu et al. 2004b; Reszka et al. 2007; Shankar et al. 2008; Faris and Friesen 2009; Friesen et al. 2009, 2012; Adhikari et al. 2011; Abeysekara et al. 2012; Ruud et al. 2017; Phan et al. 2018), on 1A, 1B, 2A, 2D, 3A, 3B, 4B, 5A, 5B, 7B and 7A for adult plant leaf resistance (Aguilar et al. 2005; Shankar et al. 2008; Friesen et al. 2009; Francki et al. 2011; Lu and Lillemo 2014; Jighly et al. 2016; Ruud et al. 2017; Francki et al. 2018), and on 2A, 2B, 2D, 3A, 3B, 4A, 4B, 5A, 5B, 6B, 7A and 7D for glume resistance (Schnurbusch et al. 2003; Aguilar et al. 2005; Uphaus et al. 2007; Shankar et al. 2008; Shatalina et al. 2014; Jighly et al. 2016; Francki et al. 2018).
In recent years, necrotrophic effectors (NEs, also known as host-selective toxins) involved in the SNB have been identified (reviewed by Friesen and Faris 2010; Oliver et al. 2012). These NEs are defined as pathogen small protein molecules that induce toxicity and promote disease. The wheat–P. nodorum pathosystem involves multiple NEs that are recognized by corresponding dominant host sensitivity genes in an inverse gene-for-gene manner to confer susceptibility to the pathogen (Oliver et al. 2012). As the pathogen produces an effector that promotes disease and the host produces a receptor that is required for susceptibility, these NE-based systems are a mirror image of the classical gene-for-gene systems often found in biotrophic systems (Wolpert et al. 2002). To date, nine host gene–necrotrophic effector interactions that play a significant role in SNB disease development have been identified: Tsn1-SnToxA (Liu et al. 2006), Snn1-SnTox1 (Liu et al. 2004a, b), Snn2-SnTox2 (Friesen et al. 2007), Snn3-B1-SnTox3 (Friesen et al. 2008b), Snn3-D1-SnTox3 (Zhang et al. 2011), Snn4-SnTox4 (Abeysekara et al. 2009), Snn5-SnTox5 (Friesen et al. 2012), Snn6-SnTox6 (Gao et al. 2015) and Snn7-SnTox7 (Shi et al. 2015). Most of these interactions show large additive effects when multiple interactions are expressed, but some show epistatic effects as well, which may explain the quantitative nature of SNB resistance (Friesen et al. 2007, 2008b; Chu et al. 2010). In addition, the disease level can be influenced by various levels of NE expression in different isolates (Faris et al. 2011) or suppressed by each other (Phan et al. 2016). Some of the reported QTL were identified in regions of chromosomes 1AS, 1BS, 2DS, 4BL, 5BS, and 5BL that correspond to the necrotrophic effector sensitivity loci. However, for other identified resistance QTL such co-location was not observed, as reviewed by Francki (2013). Interesting data on the role of Snn-SnTox interactions for SNB adult plant resistance were provided by studies of Friesen et al. (2009) and Ruud et al. (2017). Significant SnToxA-Tsn1 and SnTox2-Snn2 interaction was detected and accounted for as much as 18% and 15% of the variation in disease severity on the flag leaf, respectively, after inoculation with a single isolate (Friesen et al. 2009). QTL analysis of adult plant SNB resistance under natural pathogen infection revealed a significant effect of the SnTox3-Snn3 interaction that could explain up to 24% of the phenotypic variation (Ruud et al. 2017).
A DH mapping population of cvs. Liwilla × Begra was used in a previous study to identify QTL associated with resistance to P. nodorum at the seedling stage (Czembor et al. 2003). In this study we were interested in the SNB reaction for the same population and parents at the adult plant stage.
Materials and methods
Plant material and fungal isolates
We used a set of 74 winter wheat doubled-haploid (DH) lines that were produced in a previous study (Czembor et al. 2003) from anther cultures of the F1 hybrids derived from a cross between the SNB resistant cv. Liwilla and susceptible cv. Begra. The inoculum was produced according to the procedure previously described by Czembor et al. (2003). Artificial inoculation was conducted using a mixture of 12–15 P. nodorum isolates from different geographic regions of Poland, the same as used in a previous study (Czembor et al. 2003). In the years 2006–2008, 12 such isolates were available. The suspension of spores was adjusted to the concentration of 1.5 × 106 to 8 × 106 spores/ml and supplemented with a few drops of a surfactant (polyoxyethylene–sorbitan monolaurate, Sigma-Aldrich, Poznań, Poland) before inoculation (100 ml per 1 m2).
Adult plant testing for SNB resistance
Pathology tests were performed for adult plants under two contrasting environments: field (in the years 2002, 2003 and 2006) and polytunnel (in the years 2007 and 2008) (Table 1). In field experiments, DH lines and parents were sown into two plots arranged in two parallel mirror image lanes with two replications. Plants in one lane were inoculated at three different BBCH (Biologische Bundesanstalt, Bundessortenamt und CHemische Industrie) growth stages (Hack et al. 1992): 37 (flag leaf just visible), 59 (end of heading) and 65/69 (full flowering/end of flowering). Every inoculation was in the evening so that moisture was retained on the leaf surface overnight to promote infection. At the same time, the plants in the second parallel (mirror) lane were protected with the fungicide Tilt (0.1% propiconazole) as controls. The SNB assessment of plants was started after the appearance of the first symptoms. A 9-digit scale (1 = resistant, 9 = susceptible) was used to evaluate disease severity. The disease ratings were repeated at weekly intervals until the senescence of plants. A total of four disease readings were made on leaves and heads per season to assess Septoria nodorum leaf blotch (SNL) and Septoria nodorum glume blotch (SNG), respectively. The SNB severity on leaves and heads was expressed as the areas under the disease progress curves (AUDPC) (Shaner and Finney 1977).
In the polytunnel experiments, the mapping population and the parents were sown in 1 m long rows spaced at 18 cm in three randomized blocks, of which two were inoculated and one served as an untreated control. Plants were inoculated twice at BBCH growth stage 59 (end of heading and flag leaf fully emerged) with a two-day interval. To promote infection and symptom development plants were sprayed with water twice a day from sprinklers mounted above plots. The polytunnel prevented potential leaf and glum infections that could be caused by other pathogens. Fourteen days after the first inoculation, eight to ten leaves and heads from each line per replicate were collected, mounted on self-adhesive foil, and photographed using a digital camera. The images were analysed using the WinCam software (Regent Instruments Inc. 2004) to determine the proportion of the leaf and glume area covered by necrosis. In both experiments, in the field and the polytunnel, heading dates and heights of plants were noted.
Statistical analysis
The distribution frequency was determined for disease parameters (either AUDPC or percentage of necrotic area) observed for leaves and heads. All phenotypic data were subjected to analysis of variance (ANOVA) using the PROC GLM procedure (SAS Institute, Inc. 2009). The broad-sense heritability (h2) was estimated (Mądry et al. 2010) for all disease parameters, as well as plant height and the heading date. For each experiment, Pearson’s correlation coefficients were estimated for the relationships between plant height, heading dates and either AUDPC or the percentage of necrotic area on leaf/head.
Genetic map
The previously constructed genetic map for the Liwilla × Begra population (Czembor et al. 2007) was used for the QTL analysis. This map is far from perfect, mainly due to low DNA polymorphism between parents and the small size of the mapping population. The map comprises 235 simple sequence repeat (SSR) and diversity arrays technology (DArT) markers assigned to 18 linkage groups (lack of linkage groups for chromosomes 1A, 7B and 4D), resulting in a map of 1705 cM total length. A less complete map was observed for chromosomes in the D genome. In order to determine linkage or pleiotropy between wheat phenological traits (plant height and heading date) and SNB resistance after the first round of interval mapping (IM), additional molecular markers linked to vernalization (VRN), photoperiod (PPD) loci and plant height reducing (dwarfing) Rht genes were tested: VRN-A1 on chromosome 5A, PCR primers VrnN_FP3/VrnN_RP3 (Whittal et al. 2018); PPD-D1 on 2D, PCR primers Ppd-D1_F1/ Ppd-D1_R1/ Ppd-D1_R2 (Whittal et al. 2018); Rht8 on 2D, SSR marker wmc503 (KOMUGI 2018); Rht9 on 5A, barc151 (Ellis et al. 2005); Rht12 on 5A, gwm291 (KOMUGI 2018) and wmc410 (Ellis et al. 2005). New markers were included in the right chromosome map using JoinMap 4.0 (Van Ooijen 2006) with default settings. It is worth noting that markers linked to Rht-A1a on 4A (gwm610) and Rht22 (gwm350 proximal flanking marker on 7A) were already localized on the map (KOMUGI 2018).
QTL analysis
For the QTL analysis, 20 data sets were used to represent combinations of five years of experiments and four analysed traits (Table 1). QTL analyses were performed with MapQTL 6.0 (van Ooijen 2009) using a regression model and basically followed the strategy described by Lu and Lillemo (2014). In the QTL analyses of disease parameters, the corresponding traits plant height and heading date from the same year of the experiment were used as covariates. In the first step, interval mapping identified markers with the highest limit of detection (LOD) values by using the automatic cofactor selection option. These markers were used as cofactors for multiple-QTL mapping (MQM). The LOD threshold value for each QTL was calculated from 1000 permutation tests at the 0.05 significance level. Genetic maps of chromosomes and the detected QTL were drawn using the MapChart 2.2 software (Voorrips 2002). The confidence intervals for the QTL effects were established using the one-LOD rule (Conneally et al. 1985; Lander and Botstein 1989); these intervals are indicated in the figures as boxes. To increase the likelihood that the interval would contain the QTL, a support interval calculated as a two-LOD rule is indicated by the lines shown in the figures.
Results
Analysis of disease parameters
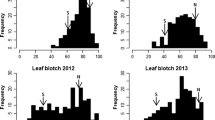
A relatively broad range of variation was found among the DH progeny and parents for the measured disease parameters (leaf and glume infection), as indicated by the coefficient of variation ranging from 12.51 to 41.23% (Table 2). The analysis of variance showed significant differences between the genotypes in all years of experiments. The population showed a continuous distribution, and parents, i.e. Liwilla and Begra, were always on the opposite sides of the scale, representing resistance and susceptibility, respectively (Table 2, Fig. 1). However, differences in reactions to SNB for parents were definitely less pronounced on heads (glumes) than on leaves, especially for the polytunnel experiments (t07nec_h and t08nec_h, Fig. 2). The susceptible parent Begra showed extremely high values for either the AUDPC on leaves or the percentage of necrotic leaf area in only three data sets: f06audpc_l, t07nec_l and t08nec_l (Table 2). In the year 2002 only (data set f02audpc_l), the resistant parent Liwilla was observed among DH lines in the class with extremely low AUDPC for leaves (Table 2). In the population, some of the offspring genotypes had higher or lower values than the resistant or susceptible parent, respectively (Table 2), indicating transgressive segregation for leaf and glume resistance.
Frequency distributions of disease parameters in the Liwilla × Begra DH population. The data set designation is shown in the upper right corner of each histogram and is explained in Table 1. The average values for resistant parent Liwilla (L) and susceptible parent Begra (B) are indicated by arrows
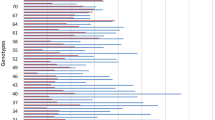
Location of QTL for resistance to Stagonospora nodorum blotch on leaves (red filled boxes) and heads (green) and for plant height (dark blue), and for heading date (bright blue) in a Liwilla × Begra doubled-haploid population. The QTL were detected using MQM mapping on chromosomes 3A, 4A, 5A, 7A, 1B, 2D and 7D. Genetic distances are shown in centimorgans (cM) to the right of each linkage group, whereas the name of the identified QTL is shown to the left. The length of the vertical boxes indicates the one-LOD confidence interval, whereas the two-LOD confidence interval is indicated by a line for the QTL detected using a given data set. The map of each chromosome was published previously by Czembor et al. (2007)
Analysis of developmental and morphological traits
A continuous distribution for plant height and heading date (data not shown) and ANOVA showed significant differences between genotypes (Table 2). However, the coefficient of variation was low for plant height (6.84%–11.32%) and even lower for heading date (1.01%–1.90%). The resistant parent Liwilla was always taller than the susceptible parent Begra, and both had the same or similar heading dates (Table 2). Among the offspring more extreme values were observed, indicating transgressive segregation for both traits (Table 2).
Heritabilities and correlations
Moderate and high heritability (Table 2) was observed for disease parameters (0.60–0.92), plant height (0.71–0.95) and heading date (0.82–0.92), indicating that the phenotypic data of the DH lines was suitable for QTL analysis. A negative correlation between disease parameters and either plant height or heading date was observed; in most cases it was either weak or moderate but statistically significant (Table 3). Only in the year 2008 were necrotic areas on leaves (t08nec_l) and heads (t08nec_h) not significantly correlated with plant height. The only strong correlations were found between heading date and AUDPC on heads in 2006 (−0.72) and necrotic area on heads in 2007 (−0.78). There was a moderate positive and significant correlation (0.38–0.67) between disease severity on heads and leaves in all years of experiments (Table 3).
QTL analysis for SNB resistance
The QTL analysis of 10 data sets (combination of the two disease resistance traits and years of the experiment) revealed five QTL associated with resistance to SNB (Table 4, Fig. 2): two with SNL resistance (QSnl.ihar-1B and QSnl.ihar-7D) and three associated with SNG resistance (QSng.ihar-3A.1, QSng.ihar-3A.2 and QSng.ihar-4A). The phenotypic variation explained by individual SNG resistance QTL ranged from 9.1 to 10.3% and was detected only once across different years (Table 4). The SNG resistance was contributed by cv. Liwilla for the QTL on chromosome 3A and by the susceptible parent cv. Begra for the QTL on 4A. Similarly, only a small proportion of the total phenotypic variation was explained by individual SNL resistance QTL, all contributed by the Begra allele, and ranging from 11.4 to 20.0% (Table 4). The QTL mapped on chromosome 7D (QSnl.ihar-7D) had the largest effect among identified loci and explained up to 20.0% of the phenotypic variance and was detected in the years 2003 and 2006. In the case of SNL resistance, a QTL was detected only once on 1B (QSnl.ihar-1B) and explained 12.4% of the phenotypic variance (Table 4). No overlapping position of individual SNL and SNG resistance QTL was observed.
QTL analysis for plant height and heading date
Among additional molecular markers linked to wheat phenological traits, no DNA polymorphism was observed for vernalization locus VRN-A1 on chromosome 5A. The other tested markers were polymorphic and the following loci were localized on the genetic map: PpdD1 and Rht8 on 2D (marker wmc503), Rht9 on 5A (barc151) and Rht12 on 5A (gwm291 and wmc410).
QTL analysis of 10 data sets (combination of plant height, heading date and year of the experiment) revealed four QTL associated with taller and later maturing plants (Table 4, Fig. 2). Two QTL were associated with heading date (QHd.ihar-2D and QHd.ihar-5A), and two with plant height (QHt.ihar-5A.2 and QHt.ihar-7A.2). The phenotypic variation explained by individual QTL for the heading date ranged from 16.7 to 32.9% (Table 4). From two QTL associated with heading date, QHd.ihar-2D was detected four times (in the years 2002, 2006, 2007 and 2008), with the highest LOD values corresponding to the PpdD1 gene, and was contributed by the Liwilla allele promoting later maturing plants. The second QTL QHd.ihar-5A on chromosome 5A was detected only once, explained 19.4% of phenotypic variance and was contributed by the Begra allele. Two QTL associated with plant height, QHt.ihar-5A.2 and QHt.ihar-7A.2, were detected in the conducted experiments one and three times, respectively, and each of them could explain 14.1–29.8% of the phenotypic variance (Table 4). The QTL QHt.ihar-7A.2 was contributed by the Liwilla allele and QHt.ihar-5A.2 by the Begra allele, both related to taller plants.
Discussion
Winter wheat cultivars Liwilla and Begra are known to differ substantially in reaction to SNB at the seedling stage, and resistance is controlled at least by several QTL (Czembor et al. 2003). Therefore, it was interesting to investigate next the reaction of the DH population Liwilla × Begra at the adult plant stage. Resistance to SNG and SNL showed a continuous phenotypic distribution reflecting their quantitative nature. Moreover, five QTL associated with resistance to SNB were identified, indicating polygenic inheritance, which is in agreement with several previous studies on adult plant resistance in wheat (Schnurbusch et al. 2003; Aguilar et al. 2005; Uphaus et al. 2007; Shankar et al. 2008; Francki et al. 2011; Lu and Lillemo 2014; Ruud et al. 2017; Francki et al. 2018). None of the DH lines were fully resistant on glumes and leaves, but more frequently they were less diseased than the resistant parent Liwilla (transgressive segregation observed), and alleles from the susceptible parent Begra also contributed to the resistance level. These findings have also been documented by other authors (Schnurbusch et al. 2003; Aguilar et al. 2005; Uphaus et al. 2007; Shankar et al. 2008; Lu and Lillemo 2014; Ruud et al. 2017; Francki et al. 2018).
QTL analysis of the adult plant SNB severity in the Liwilla × Begra mapping population revealed two QTL with SNL resistance (QSnl.ihar-1B and QSnl.ihar-7D) and three were associated with SNG resistance (QSng.ihar-3A.1, QSng.ihar-3A.2 and QSng.ihar-4A). Each of the detected QTL explained only a small proportion of the total phenotypic variation, ranging from 9.1 to 20.0%, similar to previous reports for adult plant resistance (Schnurbusch et al. 2003; Aguilar et al. 2005; Uphaus et al. 2007; Shankar et al. 2008; Lu and Lillemo 2014; Ruud et al. 2017; Francki et al. 2018). Resistance to SNL was moderately correlated with SNG (0.38–0.67) without a common region for detected QTL (Table 4, Fig. 2). Therefore, our study indicates that resistance to SNL and SNG is independent and controlled by different loci, as suggested by previous studies (Fried and Meister 1987; Bostwick et al. 1993; Wicki et al. 1999) and reports combining QTL analysis and molecular markers (Aguilar et al. 2005; Shankar et al. 2008).
On chromosome 3A, two QTL for SNG resistance contributed by the Liwilla allele were localized on opposite ends of the chromosome, i.e. QSng.ihar-3A.1 (marker internal wPt6012─wPt3041 on 3AS) and QSng.ihar-3A.2 (peaked at markers wPt9049 and wPt2698 on 3AL) accounted for 9.5% and 10.3% of the observed variation, respectively (Table 4 and Fig. 2). The same chromosome was reported to be involved in SNB adult plant resistance in previous studies (Schnurbusch et al. 2003; Aguilar et al. 2005; Lu and Lillemo 2014; Ruud et al. 2017). A recombinant inbred line (RIL) population derived from a cross between winter wheat cv. Forno and winter spelt (Triticum spelta L.) cv. Oberkulmer was evaluated for resistance to SNB on leaves and glumes for 2 years after artificial inoculation with P. nodorum (Aguilar et al. 2005). The authors identified on chromosome 3A two minor QTL for SNG resistance, QSng.eth-3A1 (R2 = 0.3%, contributed by the Oberkulmer allele) and QSng.eth-3A2 (R2 = 6.4%, contributed by the Forno allele), in the marker intervals Xpsr304─Xpsr598 and Xglk118─Xglk577, respectively. Comparative analysis of genetic maps (GrainGenes 2018, CMap tool; Quraishi et al. 2017, high resolution wheat consensus map; Diversity Arrays Technology 2018, consensus map of wheat v. 4.0) does not allow one to conclusively confirm or refute their identity with our QTL, because of different markers used in the two studies. Both QTL, QSng.eth-3A1 and QSng.eth-3A2, overlapped with one or a few morphological and developmental traits such as ear emergence, flowering date, culm length or ear length, so these QTL probably do not trigger a defence reaction but act as mechanical barriers or escape mechanisms (Aguilar et al. 2005). By contrast, the two QTL QSng.ihar-3A.1 and QSng.ihar-3A.2 detected in our study were not associated with any morphological or developmental traits, and therefore these QTL may have a real contribution to SNB resistance. Schnurbusch et al. (2003) studied an Arina × Forno population to identify and map QTL for SNG resistance under natural infection over 2 years in three environments. Among QTL detected, QSng.sfr-3AS (R2 = 7.1%, contributed by the Arina allele) of minor importance was flanked by markers Xcfd79a and Xgwm369. On the high resolution wheat consensus map (Quraishi et al. 2017), the next marker distal to Xcfd79a is Xwmc532 at position 25.9 cM, and the marker proximally flanking QSng.ihar-3A.1 (detected in our study) is wPt3041 at position 16.6 cM. Therefore, between these QTL positions an approximately 10 cM gap appears and may represent different SNG resistance loci. Lu and Lillemo (2014) studied leaf blotch resistance in an RIL population from a cross between SHA3/CBRD and the susceptible German spring cultivar Naxos over 4 years under field conditions with natural infection of P. nodorum supplied with mist irrigation. The genetic map was constructed using 554 SSR and DArT markers and was further saturated with the Illumina 90 K SNP chip (Ruud et al. 2017). In both studies, in the QTL analysis the same data set from field experiments was used (Lu and Lillemo 2014; Ruud et al. 2017). On the high resolution wheat consensus map (Quraishi et al. 2017), QTL for SNL resistance 3AS.1 identified by Ruud et al. (2017) (closest marker Xgwm2), which was also previously identified as 3AS by Lu and Lillemo (2014), and 3AS.2 (markers Ku_c41007_116 and Excalibur_c52446_519, only in the study of Ruud et al. (2017)) are at positions 65.0 cM and 78.4 cM, respectively, whereas the marker proximally flanking QSng.ihar-3A.1 (detected in our study) is wPt3041 at position 16.6 cM and QSng.ihar-3A.2 peaked at markers wPt9049 and wPt2698 at position 95.0 cM (Quraishi et al. 2017). Therefore, the two QTL for SNG resistance detected in our study on 3A are different from those reported by Lu and Lillemo (2014) and Ruud et al. (2017).
Close to the centromere, on the long arm of chromosome 4A, another SNG resistance QTL, QSng.ihar-4A, was detected in our study. This minor QTL (R2 = 9.1%) is contributed by the susceptible parent Begra and delimited by markers Xgwm610 and Xbarc5.2 (Table 4 and Fig. 2). In the aforementioned study by Aguilar et al. (2005), chromosome 4A was found to contain two minor QTL for SNG resistance, one on the end of the long arm (QSng.eth-4A2) and the other one, QSng.eth-4A1 (R2 = 1.4%), close to the centromere flanked by markers CD16.2 and Xglk331. Based on comparative analysis of genetic maps (GrainGenes 2018; CMap tool), the latter QTL may have the same position or be in close linkage to QSng.ihar-4A detected in our study. However, QSng.eth-4A1 overlapped with morphological and developmental traits such as ear emergence, flowering date, culm length or ear length, so probably it does not contribute to genetically controlled SNB resistance.
This study identified QTL contributed by the Begra allele for leaf resistance QSnl.ihar-1B (R2 = 12.4%) on the long arm of chromosome 1B with the highest LOD peak at marker wPt7160 and flanked proximally by wPt0705 (Table 4 and Fig. 2). Aguilar et al. (2005) identified two QTL, QSng.eth-1BS (marker interval Xglk301─Xglk317) and QSnl.eth-1BS2 (Xgwm18─Xglk483) and overlapping with culm length trait), each of them explaining ca. 7% of the phenotypic variance for resistance. Based on comparative analysis of genetic maps (Akbari et al. 2006; GrainGenes 2018, CMap tool), the position of these QTL is different from the one identified in our study. Friesen et al. (2009) evaluated an RIL mapping population BR34 × Grandin to SNL resistance under field conditions in 2 years using artificial inoculation with a fungal isolate known to produce SnToxA and SnTox2. QTL analysis revealed on chromosome 1BS SNL resistance QTL QSnb.fcu-1BS flanked by markers Xfcp267 and Xbarc240. According to the high resolution wheat consensus map (Quraishi et al. 2017), this marker interval is at least 10 cM away from the nearest marker wPt0705 proximally flanking QSnl.ihar-1B detected in our study, indicating different localization, QSnb.fcu-1BS, reported by Friesen et al. (2009). The same holds for SNL resistance QTL mapped in the population SHA3/CBRD × Naxos on 1BS (in the same region reported by Friesen et al. (2009)) with the closest marker Xwmc619 (Lu and Lillemo 2014) and SCM9 (Ruud et al. 2017). Francki et al. (2018) used an Illumina 90 K SNP chip to genotype the populations used in previous studies EGABlanco × Millewa (Francki et al. 2011), 6HRWSN125 × WAWHT2074 (Shankar et al. 2008) and P92201D5 × P91193D1 (Uphaus et al. 2007), but new high-density integrated SNP, SSR and DArT genetic maps used in QTL analysis did not identify any new QTL for SNB resistance but discriminated previously identified co-located QTL into separate but closely linked QTL. This approach resulted in localization of two overlapping SNL resistance QTL, QSnl07.daw-1B and QSnl08.daw-1B (both contributed by EGABlanco alleles) on 1BS and according to consensus map of wheat v. 4.0 (Diversity Arrays Technology 2018) ca. 100 cM away from QSnl.ihar-1B detected in our study.
In our study, the strongest QTL and the only one observed twice is QSnl.ihar-7D on the long arm of chromosome 7D, which explained up to 20% of the Septoria leaf blotch resistance (Table 4 and Fig. 2). This QTL contributed by the Begra allele is delimited by markers Xgdm46 and Xwmc157, localized at positions 183.1 cM and 234.3 cM, respectively, on the high resolution wheat consensus map (Quraishi et al. 2017). This chromosome was reported to contain two QTL for SNG resistance in a genome-wide association study (GWAS) on a set of synthetic hexaploid wheat (Jighly et al. 2016) mapped at a position close to 35 cM and 125 cM, respectively, based on comparative analysis of a high resolution wheat consensus map (Quraishi et al. 2017) and consensus map of wheat v. 4.0 (Diversity Arrays Technology 2018). Therefore, QSnl.ihar-7D detected in our study resides at a different position on chromosome 7D compared to those reported by Jighly et al. (2016).
Variation in morphological and developmental traits can greatly affect QTL analysis in making appropriate conclusions on loci responsible for genetically triggered defence mechanisms or those related to plant disease escape. Previous studies of Fried and Meister (1987) and Bostwick et al. (1993) demonstrated a significant correlation between plant height and SNG resistance of wheat, whereas Scott et al. (1982) and Wicki et al. (1999) reported a relationship between glume resistance and late-maturing genotypes. Thus it is extremely important to dissect only those loci that truly are involved in the plant defence response if breeding programmes could efficiently apply the results of QTL disease resistance mapping research. In the present study, a negative correlation between disease parameters and morphological traits was observed, and in most cases it was weak or moderate (Table 3). In order to distinguish phenotypic variation contributed by plant resistance from that related to disease escape mechanisms, in the QTL analysis of disease parameters the corresponding traits plant height and heading date from the same year of the experiment were used as covariates (Lu and Lillemo 2014). This statistical approach resulted in identification of no overlapping SNB resistance QTL with those related to plant morphological and developmental traits, providing more accurate and reliable information on each QTL (Table 4 and Fig. 2). In this study, two QTL were associated with heading date (QHd.ihar-2D and QHd.ihar-5A), and two with plant height (QHt.ihar-5A.2 and QHt.ihar-7A.2). Only the QTL for heading date QHd.ihar-2D perfectly matched the gene Ppd-D1 on chromosome 2D (Table 4 and Fig. 2) responsible for plant photoperiod response (Beales et al. 2007). The other two chromosomes, 5A and 7A, were previously reported for the same population but tested for Septoria tritici blotch resistance, where identical or very close marker intervals on these chromosomes also delimited QTL associated with plant height and heading date (Radecka-Janusik and Czembor 2014). Other markers linked to plant height reducing (dwarfing) Rht genes were not associated with any morphological or developmental QTL detected in our study.
The same mapping population of Liwilla × Begra was previously evaluated for partial resistance components to SNB at the seedling stage, and four QTL were detected on chromosomes 2B, 3B, 5B and 5D (Czembor et al. 2003). None of these QTL co-located with QTL for leaf or glume blotch resistance at the adult plant stage. This is in agreement with previous studies showing that independent genes control inheritance of resistance in seedling, glumes, and flag leaves in some wheat accessions (Rosielle and Brown 1980; Fried and Meister 1987; Bostwick et al. 1993). Furthermore, the same conclusion was drawn by Shankar et al. (2008) in QTL analysis of SNB resistance at the seedling stage and adult plant stage for leaves and glumes for the DH mapping population 6HRWSN125 × WAWHT2074. None of the QTL detected at the seedling stage aligned in their chromosomal positions to QTL associated with SNG or SNL resistance (Shankar et al. 2008).
Chromosomal locations of QTL for SNL and/or SNG resistance identified in this study did not correspond to any of the P. nodorum necrotrophic effector sensitivity loci described so far (Liu et al. 2004a, 2004b, 2006; Friesen et al. 2007, 2008b, 2012; Abeysekara et al. 2009; Zhang et al. 2011; Gao et al. 2015; Shi et al. 2015). This finding is supported by studies of other authors, because most of the reported SNB resistance QTL for adult plants were not related to known necrotrophic effector sensitivity loci or some inconsistencies were observed (Shankar et al. 2008; Francki et al. 2011; Lu and Lillemo 2014). However, Friesen et al. (2009) provided evidence on susceptibility of adult plants to SNB conferred by necrotrophic effectors Tsn1 and Snn2, which were also responsible for susceptibility at the seedling stage. Both Tsn1 and Snn2 accounted for a lower magnitude of variation in SNB on adult plants compared to seedlings. Also Ruud et al. (2017) demonstrated the importance of necrotrophic effectors in SNB susceptibility, since SnTox3-Snn3 interaction in the SHA3/CBRD × Naxos population could explain up to 24% of the phenotypic variation in the field and more than 51% at the seedling stage. Both studies explain variation in necrotrophic effectors’ effects on plant susceptibility at different stages due to a plant age effect (adult plants less vulnerable to host-selective toxin activity), the impact of other diseases, and environmental effects being much more variable under field conditions (Friesen et al. 2009; Ruud et al. 2017). These effects might explain why in the Liwilla × Begra population tested in this study at the adult plant stage for SNB severity under field and polytunnel conditions we did not detect the Tsn1 necrotrophic effector sensitivity locus which corresponded to QSnl.ihar-5B, a major QTL identified previously at the seedling stage for the same mapping population (Czembor et al. 2003; Francki 2013). Furthermore, in the European population of P. nodorum (including Poland) isolates produce necrotrophic effectors Tox1 and Tox3 in a frequency of 70–90%, but ToxA only at ca. 10% (McDonald et al. 2013; Walczewski and Arseniuk 2015). We cannot exclude the possibility that under field conditions due to environmental variability (Friesen et al. 2009; Ruud et al. 2018), plant infection by isolates of the pathogen other than those used in artificial inoculation and low representation of ToxA among the other necrotrophic effectors, Tsn1–SnToxA interaction might not be detected.
The present study underlines the importance of many variables that may have an impact on research on identifying genetic factors associated with SNB resistance in wheat. Several QTL detected in our study, i.e. QSng.ihar-3A.1, QSng.ihar-3A.2, QSng.ihar-4A, QSnl.ihar-1B and QSnl.ihar-7D, may represent a novel minor SNB resistance QTL contributed by both parental alleles. However, only QSnl.ihar-7D was detected twice, but none of them were co-localized with QTL associated with morphological and developmental traits, and thus could be involved in the defence reaction. Only a small portion of the phenotypic variation for SNB resistance was explained in the QTL analysis. A few possible reasons may be responsible for the results obtained, including low map resolution (some regions harbouring resistance loci might have been omitted from the map), a small mapping population, and a large effect of the genotype × environment interactions, especially as the SNB severity was evaluated under different environments of field and partially controlled polytunnel conditions. In future experiments it would be advisable to use single isolates of the pathogen with known toxin activity.
References
Abeysekara, N. S., Friesen, T. L., Keller, B., & Faris, J. D. (2009). Identification and characterization of a novel host-toxin interaction in the wheat-Stagonospora nodorum pathosystem. Theoretical and Applied Genetics, 120, 117–126.
Abeysekara, N. S., Faris, J. D., Chao, S., McClean, P. E., & Friesen, T. L. (2012). Whole-genome QTL analysis of Stagonospora nodorum blotch resistance and validation of the SnTox4-Snn4 interaction in hexaploid wheat. Phytopathology, 102, 94–104.
Adhikari, T. B., Jackson, E. W., Gurung, S., Hansen, J. M., & Bonman, J. M. (2011). Association mapping of quantitative resistance to Phaeosphaeria nodorum in spring wheat landraces from the USDA National Small Grains Collection. Phytopathology, 101, 1301–1310.
Aguilar, V., Stamp. P., Winzeler, M., Winzeler, H., Schachermayr, G., Keller, B., … Messmer, M. M. (2005). Inheritance of field resistance to Stagonospora nodorum leaf and glume blotch and correlations with other morphological traits in hexaploid wheat (Triticum aestivum L.). Theoretical and Applied Genetics, 111, 325–336.
Akbari, M., Wenzl, P., Caig, V., Carling, J., Xia, L., Yang, S., et al. (2006). Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theoretical and Applied Genetics, 113, 1409–1420.
Arseniuk, E., Góral, T., & Scharen, A. L. (1998). Seasonal patterns of spore dispersal of Phaeosphaeria spp. and Stagonospora spp. Plant Disease, 82, 187–194.
Arseniuk, E., Czembor, P. C., Czaplicki, A., Song, Q. J., Cregan, P. B., Hoffman, D. L., & Ueng, P. P. (2004). QTL controlling partial resistance to Stagonospora nodorum leaf blotch in winter wheat cultivar Alba. Euphytica, 137, 225–231.
Bathgate, J. A., & Loughman, R. (2001). Ascospores are a source of inoculum of Phaeosphaeria nodorum, P. avenaria f. sp. avenaria and Mycosphaerella graminicola in Western Australia. Australasian Plant Pathology, 30, 317–322.
Beales, J., Turner, A., Griffiths, S., Snape, J., & Laurie, D. A. (2007). A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theoretical and Applied Genetics, 115, 721–733.
Bhathal, J. S., Loughman, R., & Speijers, J. (2003). Yield reduction in wheat in relation to leaf disease from yellow (tan) spot and septoria nodorum blotch. European Journal of Plant Pathology, 109, 435–443.
Blixt, E., Djurle, A., Yuen, J., & Olson, Å. (2009). Fungicide sensitivity in Swedish isolates of Phaeosphaeria nodorum. Plant Pathology, 58, 655–664.
Bostwick, D. E., Ohm, H. W., & Shaner, G. (1993). Inheritance of septoria glume blotch resistance in wheat. Crop Science, 33, 439–443.
Chu, C. G., Faris, J. D., Xu, S. S., & Friesen, T. L. (2010). Genetic analysis of disease susceptibility contributed by the compatible Tsn1-SnToxA and Snn2-SnTox1 interactions in the wheat-Stagonospora nodorum pathosystem. Theoretical and Applied Genetics, 120, 1451–1459.
Conneally, P. M., Edwards, J. H., Kidd, K. K., Lalouel, J.-M., Morton, N. E., Ott, J., & White, R. (1985). Report of the committee and methods of linkage analysis and reporting. Cytogenetics and Cell Genetics, 40, 356–359.
Czembor, P. C., Arseniuk, E., Czaplicki, A., Song, Q. J., Cregan, P. B., & Ueng, P. P. (2003). QTL mapping of partial resistance in winter wheat to Stagonospora nodorum blotch. Genome, 46, 546–554.
Czembor, P. C., Radecka, M., & Arseniuk, E. (2007). Mapa molekularna pszenicy (Triticum aestivum L.). Biuletyn IHAR, 243, 279–288.
Diversity Arrays Technology Pty. Ltd., (2018). A consensus map of wheat v. 4.0. https:/www.diversityarrays.com/technology-and-resources/genetic-maps/. Accessed 20 December 2018.
Du, C. G., Nelson, L. R., & McDaniel, M. E. (1999). Diallel analysis of gene effects conditioning resistance to Stagonospora nodorum (Berk.) in wheat. Crop Science, 39, 686–690.
Ecker, R., Dinoor, A., & Cahaner, A. (1989). The inheritance of resistance to Septoria glume blotch: I. common bread wheat, Triticum aestivum. Plant Breeding, 102, 113–121.
Ellis, M. H., Rebetzke, G. J., Azanza, F., Richards, R. A., & Spielmeyer, W. (2005). Molecular mapping of gibberellin-responsive dwarfing genes in bread wheat. Theoretical and Applied Genetics, 111, 423–430.
Eyal, Z. (1981). Integrated control of septoria diseases of wheat. Plant Disease, 65, 763–768.
Faris, J. D., & Friesen, T. L. (2009). Reevaluation of a tetraploid wheat population indicates that the Tsn1-ToxA interaction is the only factor governing Stagonospora nodorum blotch susceptibility. Phytopathology, 99, 906–912.
Faris, J. D., Zhang, Z., Rasmussen, J. B., & Friesen, T. L. (2011). Variable expression of the Stagonospora nodorum effector SnToxA among isolates is correlated with levels of disease in wheat. Molecular Plant-Microbe Interactions, 24, 1419–1426.
Francki, M. G. (2013). Improving Stagonospora nodorum resistance in wheat: A review. Crop Science, 53, 355–365.
Francki, M. G., Shankar, M., Walker, E., Loughman, R., Golzar, H., & Ohm, H. (2011). New quantitative trait loci in wheat for flag leaf resistance to Stagonospora nodorum blotch. Phytopathology, 101, 1278–1284.
Francki, M. G., Walker, E., Li, D. A., & Forrest, K. (2018). High-density SNP mapping reveals closely linked QTL for resistance to Stagonospora nodorum blotch (SNB) in flag leaf and glume of hexaploid wheat. Genome, 61, 145–149.
Fried, P. M., & Meister, E. (1987). Inheritances of leaf and head resistance of winter wheat to Septoria nodorum in diallel cross. Phytopathology, 86, 459–463.
Friesen, T. L., & Faris, J. D. (2010). Characterization of the wheat–Stagonospora nodorum disease system: What is the molecular basis of this quantitative necrotrophic disease interaction? Canadian Journal of Plant Pathology, 32, 20–28.
Friesen, T. L., Meinhardt, S. W., & Faris, J. D. (2007). The Stagonospora nodorum-wheat pathosystem involves multiple proteinaceous host-selective toxins and corresponding host sensitivity genes that interact in an inverse gene-for-gene manner. The Plant Journal, 51, 681–692.
Friesen, T. L., Faris, J. D., Solomon, P. S., & Oliver, R. P. (2008a). Host-selective toxins: Effectors of necrotrophic pathogenicity. Cellular Microbiology, 10, 1421–1428.
Friesen, T. L., Zhang, Z. C., Solomon, P. S., Oliver, R. P., & Faris, J. D. (2008b). Characterization of the interaction of a novel Stagonospora nodorum host-selective toxin with a wheat susceptibility gene. Plant Physiology, 146, 682–693.
Friesen, T. L., Chu, C. G., Liu, Z. H., Xu, S. S., Halley, S., & Faris, J. D. (2009). Host-selective toxins produced by Stagonospora nodorum confer disease susceptibility in adult wheat plants under field conditions. Theoretical and Applied Genetics, 118, 1489–1497.
Friesen, T. L., Chu, C., Xu, S. S., & Faris, J. D. (2012). SnTox5–Snn5: A novel Stagonospora nodorum effector–wheat gene interaction and its relationship with the SnToxA–Tsn1 and SnTox3–Snn3–B1 interactions. Molecular Plant Pathology, 13, 1101–1109.
Gao, Y., Faris, J. D., Liu, Z., Kim, Y. M., Syme, R. A., Oliver, R. P., Xu, S. S., & Friesen, T. L. (2015). Identification and characterization of the SnTox6-Snn6 interaction in the Parastagonospora nodorum-wheat pathosystem. Molecular Plant-Microbe Interactions, 28, 615–625.
GrainGenes. (2018) (Accessed 23 February 2018). A Database for Triticeae and Avena, https://wheat.pw.usda.gov/GG3/.
Hack, H., Bleiholder, H., Buhr, L., Meier, U., Schnock-Fricke, U., Weber, E., & Witzenberger, A. (1992). Einheitliche Codierung der phänologischen Entwicklungsstadien mono- und dikotyler Pflanzen – Erweiterte BBCH-Skala. Nachrichtenblatt des Deutschen Pflanzenschutzdienstes, 44, 265–270.
Jighly, A., Alagu, M., Makdis, F., Singh, M., Singh, S., Emebiri, L. C., & Ogbonnaya, F. (2016). Genomic regions conferring resistance to multiple fungal pathogens in synthetic hexaploid wheat. Molecular Breeding, 36, 127.
Keller, S. M., McDermott, J. M., Pettway, R. E., Wolfe, M. S., & McDonald, B. A. (1997). Gene flow and sexual reproduction in the wheat glume blotch pathogen Phaeosphaeria nodorum (anamorph Stagonospora nodorum). Phytopathology, 87, 353–358.
KOMUGI, (2018). Wheat Genetic Resources Database. https://shigen.nig.ac.jp/wheat/komugi/. Accessed 20 December 2018.
Lander, E., & Botstein, D. (1989). Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics, 121, 185–199.
Liu, Z. H., Faris, J. D., Meinhardt, S. W., Ali, S., Rasmussen, J. B., & Friesen, T. L. (2004a). Genetic and physical mapping of a gene conditioning sensitivity in wheat to a partially purified host-selective toxin produced by Stagonospora nodorum. Phytopathology, 94, 1056–1060.
Liu, Z. H., Friesen, T. L., Rasmussen, J. B., Ali, S., Meinhardt, S. W., & Faris, J. D. (2004b). Quantitative trait loci analysis and mapping of seedling resistance to Stagonospora nodorum leaf blotch in wheat. Phytopathology, 94, 1061–1067.
Liu, Z. H., Friesen, T. L., Ling, H., Meinhardt, S. W., Oliver, R. P., Rasmussen, J. B., & Faris, J. D. (2006). The Tsn1-ToxA interaction in the wheat-Stagonospora nodorum pathosystem parallels that of the wheat-tan spot system. Genome, 49, 1265–1273.
Lu, Q., & Lillemo, M. (2014). Molecular mapping of adult plant resistance to Parastagonospora nodorum leaf blotch in bread wheat lines ‘Shanghai-3/catbird’ and ‘Naxos’. Theoretical and Applied Genetics, 127, 2635–2644.
Mądry, W., Mańkowski, D. R., Kaczmarek, Z., Krajewski, P., & Studnicki, M. (2010). Metody statystyczne oparte na modelach liniowych w zastosowaniach do doświadczalnictwa, genetyki i hodowli roślin. Monografie i rozprawy naukowe IHAR, 34.
McDonald, B. A., & Linde, C. (2002). The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica, 124, 163–180.
McDonald, M. C., Oliver, R. P., Friesen, T. L., Brunner, P. C., & McDonald, B. A. (2013). Global diversity and distribution of three necrotrophic effectors in Phaeosphaeria nodorum and related species. New Phytologist, 199, 241–251.
Mullaney, E. J., Martin, J. M., & Scharen, A. L. (1982). Generation mean analysis to identify and partition the components of genetic resistance to Septoria nodorum in wheat. Euphytica, 31, 539–545.
Nelson, L. R., & Gates, C. E. (1982). Genetics of host plant resistance of wheat to Septoria nodorum. Crop Science, 22, 771–773.
Oliver, R. P., Friesen, T. L., Faris, J. D., & Solomon, P. S. (2012). Stagonospora nodorum: From pathology to genomics and host resistance. Annual Review of Phytopathology, 50, 23–43.
Phan, H. T., Rybak, K., Furuki, E., Breen, S., Solomon, P. S., Oliver, R. P., & Tan, K. C. (2016). Differential effector gene expression underpins epistasis in a plant fungal disease. The Plant Journal, 87, 343–354.
Phan, H. T. T., Rybak, K., Bertazzoni, S., Furuki, E., Dinglasan, E., Hickey, L. T., Oliver, R. P., & Tan, K.-C. (2018). Novel sources of resistance to Septoria nodorum blotch in the Vavilov wheat collection identified by genome-wide association studies. Theoretical and Applied Genetics, 131, 1223–1238.
Quaedvlieg, W., Verkley, G. J., Shin, H. D., Barreto, R. W., Alfenas, A. C., Swart, W. J., et al. (2013). Sizing up septoria. Studies in Mycology, 75, 307–390.
Quraishi, U. M., Pont, C., Ain, Q., Flores, R., Burlot, L., Alaux, M., Quesneville, H., & Salse, J. (2017). Combined genomic and genetic data integration of major agronomical traits in bread wheat (Triticum aestivum L.). Frontiers in Plant Science, 8, 1843.
Radecka-Janusik, M., & Czembor, P. C. (2014). Genetic mapping of quantitative trait loci (QTL) for resistance to septoria tritici blotch in a winter wheat cultivar Liwilla. Euphytica, 200, 109–125.
Regent Instruments, Inc. (2004). WinCam User’s guide. Version 2004a. Canada Regent Instruments Inc., Quebec, Canada.
Reszka, E., Song, Q. J., Arseniuk, E., Cregan, P. B., & Ueng, P. P. (2007). The QTL controlling partial resistance to Stagonospora nodorum blotch disease in winter triticale ‘Bogo’. Plant Pathology Bulletin, 16, 161–167.
Rosielle, A. A., & Brown, A. G. P. (1980). Selection for resistance to Septoria nodorum in wheat. Euphytica, 29, 337–346.
Rufty, R. C., Hebert, T. T., & Murphy, C. F. (1981). Evaluation of resistance to Septoria nodorum in wheat. Plant Disease, 65, 406–409.
Ruud, A. K., Dieseth, J. A., Lillemo, M. (2018) Effects of Three Necrotrophic Effectors on Spring Wheat under Norwegian Field Conditions. Crop Science 58 (1):159
Ruud, A. K., Windju, S., Belova, T., Friesen, T. L., & Lillemo, M. (2017). Mapping of SnTox3-Snn3 as a major determinant of field susceptibility Septoria nodorum leaf blotch in the SHA3/CBRD × Naxos population. Theoretical and Applied Genetics, 130, 1361–1374.
SAS Institute Inc. (2009). SAS/STAT 9.2 user’s guide. Second edition. SAS Institute Inc., Cary, NC, USA.
Schnurbusch, T., Paillard, S., Fossati, D., Messmer, M., Schachermayr, G., Winzeler, M., & Keller, B. (2003). Detection of QTLs for Stagonospora glume blotch resistance in Swiss winter wheat. Theoretical and Applied Genetics, 107, 1226–1234.
Scott, P. R., Benedikz, P. W., & Cox, C. J. (1982). A genetic study of the relationship between height, time of ear emergence and resistance to Septoria nodorum in wheat. Plant Pathology, 31, 45–60.
Shaner, G., & Buechley, G. (1995). Epidemiology of leaf blotch on soft red winter wheat caused by Septoria tritici and Stagonospora nodorum. Plant Disease, 79, 928–938.
Shaner, G., & Finney, R. E. (1977). The effect of nitrogen fertilization on the expression of slow-mildewing resistance in Knox wheat. Phytopathology, 67, 1051–1056.
Shankar, M., Walker, E., Golzar, H., Loughman, R., Wilson, R. E., & Francki, M. G. (2008). Quantitative trait loci for seedling and adult plant resistance to Stagonospora nodorum in wheat. Phytopathology, 98, 886–893.
Shatalina, M., Messmer, M., Feuillet, C., Mascher, F., Paux, E., Choulet, F., Wicker, T., & Keller, B. (2014). High-resolution analysis of a QTL for resistance to Stagonospora nodorum glume blotch in wheat reveals presence of two distinct resistance loci in the target interval. Theoretical and Applied Genetics, 127, 573–586.
Shi, G., Friesen, T. L., Saini, J., Xu, S. S., Rasmussen, J. B., & Faris, J. D. (2015). The wheat Snn7 gene confers susceptibility on recognition of the Parastagonospora nodorum necrotrophic effector SnTox7. The Plant Genome, 8, 1–10.
Solomon, P. S., Lowe, R. G. T., Tan, K.-C., Waters, O. D. C., & Oliver, R. P. (2006). Stagonospora nodorum: Cause of stagonospora nodorum blotch of wheat. Molecular Plant Pathology, 7, 147–156.
Sommerhalder, R. J., McDonald, B. A., Mascher, F., & Zhan, J. (2010). Sexual recombinants make a significant contribution to epidemics caused by the wheat pathogen Phaeosphaeria nodorum. Phytopathology, 100, 855–862.
Uphaus, J., Walker, E., Shankar, M., Golzar, H., Loughman, R., Francki, M., & Ohm, H. (2007). Quantitative trait loci identified for resistance to Stagonospora glume blotch in wheat in the USA and Australia. Crop Science, 47, 1813–1822.
Van Ooijen, J. W. (2006). JoinMap 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V., Wageningen.
Van Ooijen, J. W. (2009). MapQTL 6, software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma B.V., Wageningen.
Voorrips, R. E. (2002). MapChart: Software for the graphical presentation of linkage maps and QTLs. Journal of Heredity, 93, 77–78.
Walczewski, J., & Arseniuk, E. (2015). Produkcja białkowych toksyn przez izolaty z krajowej populacji S. nodorum. Nauka dla Hodowli i Nasiennictwa Roślin Uprawnych, Zakopane, 2.02–6.02.2015. Streszczenia Referatów i Posterów, 366.
Whittal, A., Kaviani, M., Graf, R., Humphreys, G., & Navabi, A. (2018). Allelic variation of vernalization and photoperiod response genes in a diverse set of north American high latitude winter wheat genotypes. PLoS One, 13, e0203068.
Wicki, W., Winzeler, M., Schmid, J. E., Stamp, P., & Messmer, M. (1999). Inheritance of resistance to leaf and glume blotch caused by Septoria nodorum. Theoretical and Applied Genetics, 99, 1265–1272.
Wilkinson, C. A., Murphy, J. P., & Rufty, R. C. (1990). Diallel analysis of components of partial resistance to Septoria nodorum in wheat. Plant Disease, 74, 47–50.
Wolpert, T. J., Dunkle, L. D., & Ciuffetti, L. M. (2002). Host-selective toxins and avirulence determinants: what’s in a name? Annual Review of Phytopathology, 40, 251–258.
Xu, S.S., Friesen, T.L., and Cai, X. ( 2004). Sources and genetic control of resistance to Stagonospora nodorum blotch in wheat. In S. G. Pandalai (Ed.), Recent Research Developments in Genetics and Breeding (Vol. 1 Part II., pp. 449– 469) Kerala, India: Research Signpost.
Zhang, Z. C., Friesen, T. L., Xu, S. S., Shi, G. J., Liu, Z. H., Rasmussen, J. B., & Faris, J. D. (2011). Two putatively homoeologous wheat genes mediate recognition of SnTox3 to confer effector-triggered susceptibility to Stagonospora nodorum. The Plant Journal, 65, 27–38.
Acknowledgements
The authors thank Alina Sołtys for excellent technical assistance and Dariusz Mańkowski for help with the statistical analysis.
Funding
This research was supported financially by the BIOEXPLOIT (FOOD-CT-2005-513959, Exploitation of Natural Plant Biodiversity for the Pesticide-free Production of Food) 6th European Union Framework Programme.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Research involving human participants and/or animals
The research involved neither human participants nor animals.
Informed consent
Not applicable.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Czembor, P.C., Arseniuk, E., Radecka-Janusik, M. et al. Quantitative trait loci analysis of adult plant resistance to Parastagonospora nodorum blotch in winter wheat cv. Liwilla (Triticum aestivum L.). Eur J Plant Pathol 155, 1001–1016 (2019). https://doi.org/10.1007/s10658-019-01829-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-019-01829-5