Abstract
Clubroot, caused by the protozoan parasite Plasmodiophora brassicae Woronin, is one of the most damaging diseases of Brassica napus worldwide. Resistant plant material is valuable for cultivation in all areas of high incidence of the disease and intensive growth of oilseed rape. We have evaluated clubroot resistance, plant morphology and seed quality in 15 lines of an F4 generation and selected six lines of F5 generation of interspecific hybrids obtained from a cross between a male sterile line of B. napus ‘MS8’, selected from resynthesized oilseed rape (B. rapa ssp. chinensis × B. oleracea var. gemmifera) and an ecotype of B. rapa ssp. pekinensis. Clubroot resistance was evaluated using a bioassay with P1-P5 pathotypes of P. brassicae (according to the classification of Somé et al. 1996). The resistance to the pathotype P1 was successfully fixed in the F5 generation, and improved in some lines in respect to the pathotypes P2-P4. The resistance to P1 and the other tested pathotypes was not linked. Characterization of plant material included recent techniques of FISH and BAC-FISH with a special focus on the analysis of ribosomal DNA (rDNA) of selected individuals. Two hybrid lines combined high levels of resistance with appropriate plant morphology, good seed quality traits and a stable chromosome number and arrangement. Recent techniques of ‘chromosome painting’ provided good insight into chromosome organization in the hybrids obtained, and offered opportunities of further improvement of the breeding process.
Similar content being viewed by others

Introduction
Clubroot, caused by the obligate plant pathogen Plasmodiophora brassicae Woronin, is one of the most important and commonly occurring diseases of oilseed rape, especially in Europe and North America (Robak 1991; Agrios 2005; Dixon 2009a; Lüders et al. 2011). For the last few years the disease has been an increasing concern for farmers in Poland (Korbas et al. 2009). Clubroot occurs in large areas of rapeseed cultivation, although the disease severity greatly differs between regions of the country. According to recent reports (Konieczny 2012; Jedryczka et al. 2013, 2014; Korbas et al. 2014), clubroot infestation is estimated to affect over 250,000 ha of agricultural soils in Poland, representing around one third of the acreage of oilseed rape cultivation. This outbreak is a result of the intensive cultivation of oilseed rape and lack of rotations, or only brief rotations, with non-cruciferous crop species, which is known to increase disease incidence (Robak 1994; Dixon 2009b). The pathogen (P. brassicae) is highly variable, with P1 and P3 pathotypes prevailing in Poland (Ričařová et al. 2016), according to the classification by Somé et al. (1996).
Clubroot disease development is characterized by the formation of large galls on the roots of affected plants, which hinder water and nutrient uptake and lead to yield and seed quality losses. Research on clubroot disease in Sweden has shown that infestation of about 90 % of plants resulted in a 50 % loss in seed yield (Wallenhammar et al. 1999). Grain yield losses for Polish B. rapa cultivars were 69 %, 96 % and 89 % in field trials in 1998, 1999 and 2000, respectively (Pageau et al. 2006). Very similar yield losses were observed for Argentine cultivars of B. napus (80 %- 91 %) conducted in Quebec, Canada (Pageau et al. 2006). Moreover, a significant decrease in oil content (2–6 %) and an increase in chlorophyll content in the oil were often associated with P. brassicae infection (Engqvist 1994).
The ability of P. brassicae to survive in soil as resting spores for long periods makes it difficult to control by cultural practices or chemical treatments (Voorrips 1995). Thus, breeding of resistant cultivars is a desirable means of minimizing crop losses, especially when resistance is incorporated into integrated disease management systems (Piao et al. 2009). According to Rahman et al. (2014), growing resistant cultivars in appropriate rotations is the most effective, efficient and environmentally friendly solution for the long-term management of clubroot.
Plant breeders investigate resistance in related wild species or genera and incorporate it by interspecific hybridization (Allard 1960). The backcross or pedigree methods of breeding are performed to overcome unsuitable agronomic properties of wild-type lines. With either method, one of the parents, chosen for its good agronomic characteristics, is crossed with another parent that has a high level of resistance, preferably conferred by multiple dominant genes against a wide range of clubroot pathotypes (Allard 1960; Moreno-Gonzalez and Cubero 1993). In cabbage breeding programs for disease resistance, the identification of resistance sources is performed in parallel with the recovery of marketing type and the elimination of undesirable traits from the resistance source. This is particularly difficult when inter-specific crosses are made with resistance sources (Nomura et al. 2005), or during the incorporation of the resistance trait into the desired morphotypes of B. oleracea (Bagget and Kean 1985). However, significant variability in resistance to clubroot was found among different cultivars of B. oleracea (Diederichsen et al. 2009). Resistance in B. oleracea has traditionally been considered to be non-differential, determined by a series of recessive resistance genes, and thus difficult to use in conventional breeding (Tewari and Mithen 1999; Diederichsen et al. 2009). Since the discovery and development of clubroot-resistant European turnips (Wit and Van De Weg 1964), there has been an increasing effort by researchers from different parts of the world to screen Brassica germplasm for clubroot resistance genes (CR). Among the two progenitor species of B. napus, clubroot resistance is found more frequently in turnips (B. rapa; A genome; n = 10) (Hirai 2006). Yoshikawa (1981) found CR lines in European fodder turnips and used them as sources for breeding CR Chinese cabbages. More than 50 CR F1 hybrid cultivars of Chinese cabbage have been released in Japan (Yoshikawa 1981; Kuginuki et al. 1999). However, expression of resistance is often quantitative and the genetic basis of the resistance to clubroot in B. rapa is not clear. Moreover, breakdown of disease resistance resulting from genetic variability of the pathogen has been reported (Suwabe et al. 2003; Strelkov et al. 2016). Previous experience in other countries has shown that genetic resistance can quickly break down, because the pathotype composition can shift rapidly in response to selection pressure. Changes in the population of Leptosphaeria maculans, a wind-transmitted necrotrophic ascomycete fungus causing stem canker of brassicas, have been frequently reported in oilseed rape (Li et al. 2003; Rouxel et al. 2003; Stachowiak et al. 2006; Kutcher et al. 2010; Van de Wouw et al. 2010; Kaczmarek et al. 2014).
Cultivars resistant to P. brassicae are catalogued in the Common Catalogue of Varieties of Agricultural Plant Species (CCA, European Union 2009). The first cultivar of winter oilseed rape resistant to clubroot (cv. ‘Mendel’) was a re-synthesized line of B. napus obtained from a cross between B. rapa ECD-04 × B.oleracea ECD-15, further intercrossed with the high yielding B. napus cv. ‘Falcon’ (Diederichsen and Sacristán 1996; Diederichsen et al. 2006). The same resistance source is now commonly used in other cultivars resistant to clubroot. Exploitation of a resistance gene in a resistant genotype is an approach to control the disease. It is, therefore, essential to identify the sources of resistance to clubroot.
The objective of this work was to fix the resistance to clubroot of B. rapa ssp. pekinensis accession, in plants retaining oilseed rape morphology and good seed quality in hybrids resulting from a cross with B. napus. This is the first study in which the number and rearrangement of different A and C genome chromosomes, observed using fluorescence in situ hybridization (FISH) and genomic in situ hybridization (GISH)-like techniques was implemented to achieve a deeper insight into the cytogenetic background of hybrid development.
Materials and methods
Plant material
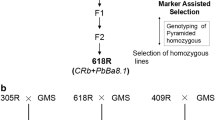
The study of an F4 generation was carried out using 15 lines of interspecific hybrids, obtained from a cross between B. napus × B. rapa ssp. pekinensis (Fig. 1). The maternal form used for obtaining hybrids was a male sterile line of an F8 generation of B. napus (MS8), selected from resynthesized oilseed rape (B. rapa ssp. chinensis × B. oleracea var. gemmifera) using in vitro cultures of isolated embryos. The maternal form B. rapa ssp. chinensis, accession number KW 171, was obtained from the Research Centre for Cultivar Testing (COBORU) located in Słupia Wielka near Poznań in 1980 and the paternal form B. oleracea var. gemmifera cultivar Maczuga (Brussels sprouts) was a Polish cultivar obtained by Produkcja i Hodowla Roślin Ogrodniczych Krzeszowice sp. z o.o. (Production and Breeding of Horticultural Plants Krzeszowice Ltd.). The genotype of B. rapa ssp. pekinensis was a local ecotype, accession number KW 786, obtained in 1978 from COBORU. All interspecific hybrid lines were sister-pollinated (five plants were placed under the same cover during flowering) for four generations in order to stabilize the fertility (Fig. 1). Earlier generations of hybrids were selected and tested for several traits such as fertility, yield, plant morphology and the uniformity of shape and size. The hybrids of the F4 generation had reasonably uniform morphological characteristics. As the maternal and paternal forms contained genotypes resistant to clubroot, this character was also studied in their progeny. The B. rapa ssp. pekinensis was fully resistant to P1 and partially resistant to the remaining pathotypes (P2-P5), with higher levels of resistance to P2 and P4 and low resistance to P3 and P5. The second round of resistance tests – done according to the identical procedure, was performed using six lines of F5 generation, selected from the plant material tested in F4 generation. The following hybrids were selected: HL05 HL06, HL07, HL08, HL10 and HL13. The selection of plants of the F4 generation, for pollination under the covers to obtain F5, was based on the results of resistance tests: lines HL05, HL06 and HL07 were selected due to very high levels of resistance to the pathotype P1, while lines HL08, HL10 and HL13 showed intermediate resistance to P1 combined to some resistance to P3. Each time the resistance test was done using the maternal and paternal forms and two standards: a susceptible B. rapa ssp. chinensis ‘Granaat’ and B. napus cv. ‘Mendel’ as a resistant control.
Evaluation of morphotypes
Morphotypes of plants of the F4 generation hybrid lines were observed and compared with the parental lines, as proposed by Wojciechowski (1993). To determine whether obtained plants were of the B. napus or B. rapa type, analysis of some selected morphological traits was performed, based on: a) leaf color (green or light-green), b) presence of trichomes on the lower side of the leaf blade (yes or no), c) position of the buds relative to the open flowers (above, in between, under), d) growth habit, e) type of inflorescence, and f) flower characters (sterile or fertile).
Resistance tests
The resistance of parental lines and intergeneric hybrids was assessed using a bioassay with P. brassicae isolates belonging to pathotypes P1-P5, as classified by the system of Somé et al. (1996). The isolates were obtained from clubroot galls found on oilseed rape plants in Poland (Table 1). The galls were chopped into small fragments and a piece of every gall was propagated in a glasshouse, in soil with the pH adjusted to 5.7 ± 0.1, on the susceptible genotype B. rapa ssp. chinensis ‘Granaat’. The screening of plants for resistance to clubroot was carried out in glasshouse conditions with a controlled temperature of 20-21 °C. The clubs were ground in distilled water with a blender, Ultra Turrax T25 Digital (IKA, Germany), and the suspension was filtered through cheese-cloth. The concentration of resting spores was determined by haemocytometer and adjusted to 1 × 106 spores/ml. Seeds of the hybrids and parental genotypes were germinated in Petri dishes for 5 days. They were planted by hand on soil and inoculated with 2 ml of spore suspension per plant. To avoid plants escaping from the infection, the inoculum was distributed by a plastic syringe; it was always injected to the soil, very close to the plant root. Seeds of the tested genotypes were sown in peat of neutral pH, mixed 2:1 with acidic peat of pH 5.5 (Biovita Ltd., Poland). There were five seeds sown to four pots (5 × 5 cm) in a potted palette, with three replicates. The assessment was carried out 6 weeks after inoculation. Before the assessment, all plants were removed from the soil, and the roots were washed for easier inspection. The evaluation of the development of the root system and general condition of the plant was assessed on a 4 point scale, where: 0 means no symptoms of the disease; 1 – stunted roots, shorter than in control plants, slightly swollen; 2 – very small clubs on some roots; 3 – big clubs, but roots still partially existing; 4 – the presence of large galls on the roots of inoculated plants, main and lateral roots entirely changed to clubs. Disease symptoms divided into 0–4 grades were used for statistical analyses. Grades 0, 1 and 2 were then jointly grouped as resistant plants (R), whereas grades 3 and 4 jointly formed the category of susceptible plants (S). The reaction of the analyzed plants was compared based on the results of statistical calculations, not only with parental forms but also with the standards. Namely, a susceptible genotype B. rapa ssp. chinensis ‘Granaat’ as well as the resistant control, B. napus cv. ‘Mendel’ (oilseed rape, winter form) were used.
Seed quality
During the growing season, two parental lines and 15 F4 hybrid lines were grown at the Poznan University of Life Sciences (PULS) experimental station Dlon, located 100 km south of Poznan. The experiment to evaluate the field performance and seed quality was done for the parental lines and 10 hybrids with the highest resistance to P. brassicae (HL02, HL03, HL05-HL08, HL10, HL12-HL14). The field study was conducted in a completely randomized block design with three replicates. The plot size was 5 m2 with 4 rows spaced 25 cm apart.
Seeds at the stage of technological ripeness from the ten most highly yielding hybrid lines and from the parental lines were harvested and analyzed for oil, protein and sinapine content. The seed samples for this analysis were collected from 20 self-pollinated plants from each tested line per plot. To determine the chemical constituents of seeds for oil content (%), protein content (%) and sinapine content (%), whole seed samples (minimum 3 g of intact seeds) were scanned on a Near Infra-Red (NIR) Spectroscopy System (6500 NIR Inc., Silverspring, MD, USA) according to the manufacturer’s protocol. The samples were scanned in triplicate to minimize sampling error.
Chromosome preparation
The study was done using the male sterile allotetraploid B. napus (MS8), diploid B. rapa ssp. pekinensis and two allotetraploid B. napus individuals of the F4 generation. Hybrids selected for the studies differed with seed quality: hybrid line HL06 had a combination of high oil and protein content (37.21 % and 22.31 %, respectively), whereas HL14 had significantly lower amounts of oil and protein (29.80 % and 20.81 %, respectively), as indicated in Table 4. Both lines contained 1.35 % of sinapine. Seeds of the selected genotypes were germinated on filter paper moistened with tap water at 20–22 °C in the dark until the roots were 1.5–2 cm long. Whole seedlings were then treated with 2 mM 8-hydroxyquinoline for 1–4 h at room temperature, fixed in a 3:1 (v/v) mixture of ethanol and glacial acetic acid, and stored at −20 °C until required. Further treatment was performed according to Hasterok et al. (2006). Chromosome analysis was carried out using an Olympus BX 60 epifluorescence microscope on 3–5 well-spread metaphase phase cells. Each chromosomal preparation was derived from a different single root tip, so that each preparation corresponded to one individual.
Fluorescence in situ hybridization (FISH)
The species-specific BoB014O06 BAC clone from a B. oleracea BAC library was used as a probe for the C-genome (GISH-like technique; Książczyk et al. 2011). The BoB014O06 clone was labelled by random priming with digoxigenin-11-dUTP (Roche). For ribosomal genes, we followed the nomenclature allowing attribution of each chromosome to a linkage group in B. rapa (Kim et al. 2009) and B. oleracea (Howell et al. 2002). In case of sites which are located on the cytogenetically undistinguishable A5, A6, and A9 chromosomes (collectively grouped as Brassica chromosomal type VIII), we followed the nomenclature proposed by Hasterok et al. (2006). The base chromosomal types, numbered I–VIII, have been introduced and described in detail by Hasterok et al. (2001), with the exception that the 5S rDNA site in chromosome type V is now assigned to the short arm (Hasterok et al. 2006). The ribosomal probes used in this study were 26S rDNA (Unfried and Gruendler 1990), used for detection of 35S rDNA loci, and pTa794 (Gerlach and Dyer 1980), which contained the 5S rDNA. The 26S rDNA was labelled with digoxigenin-11-dUTP by nick translation and pTa794 with tetramethyl-rhodamine-5-dUTP (Roche) using PCR. The FISH procedure was performed as described by Książczyk et al. (2010). Digoxigenin-labelled probes were detected with anti-digoxigenin antibody conjugated with FITC (Roche). All images were acquired using either an Olympus XM10 CCD camera attached to an Olympus BX 61 automatic epifluorescence microscope, or an F-View II CCD camera attached to an Olympus BX 60 epifluorescence microscope. Image processing and superimpositions were carried out using Olympus Cell-F imaging software and Micrografx Picture Publisher software.
Statistical calculations
Null hypothesis about the lack of differences between genotypes was verified using single factor Kruskal-Wallis test followed by post-hoc Dunn’s test for multiple comparisons (Kruskal 1952). The inference regarding the significance of differences between the seed quality was carried out on the basis of one-way analysis of variance. When analysis of variance showed no significance differences between the groups under consideration, no subsequent tests were made. If the null hypothesis was rejected, i.e., the analysis of variance showed a statistically significant difference between the seed quality, study of the differences between the means of individual groups were performed using Tukey’s test. All the reported differences and correlation coefficients were regarded as statistically significant at α ≤ 0.05. Calculations were performed according to standard procedures with Statistica 9.0 (StatSoft, Poland).
Results
Morphology of hybrid plants
Whereas the resulting plants in the F1 generation in most cases combined the characteristics of the parental genotypes, plants of the F4 generation lines were very uniform in growth habit. Morphotypes of these plants were close to oilseed rape and only in individual cases some characters were more similar to turnip rape, e.g., lighter leaf color, trichomes on the lower side of the leaf blade, and turnip rape-like inflorescence. Over 85 % of the plants were classified as ‘B. napus type-like plants’. No significant new characters, absent in either parent, were observed in the F4 hybrid lines (Fig. 2).
Resistance of parental and hybrid genotypes to clubroot
The maternal, male sterile B. napus genotype ‘MS8’ was heterogeneous in its resistance to the pathotypes P2 and P4 of P. brassicae (43 % resistant plants, that is 26 plants out of 60 tested for each of the pathotypes), but it was fully susceptible to the pathotypes P1, P3 and P5 (Table 2). The paternal genotype of B. rapa ssp. pekinensis was fully resistant to the pathotype P1 and heterogeneous in its resistance to the other pathotypes tested: P2 and P4 (60 % of resistant plants, 36 plants out of 60 tested), P5 (20 %, 12 plants out of 60 tested) and P3 (9 %, 5 plants out of 55 tested). The hybrid genotypes of the F4 generation, resulting from the cross between the parental lines, greatly differed in their resistance to particular pathotypes of P. brassicae, but it was possible to select numerous individual plants resistant to pathotypes P1-P5. The highest resistance was observed in the case of the pathotype P1, where 12 of 15 hybrid genotypes showed a resistant reaction. The highest resistance was found in line HL06 (83.3 % resistant plants, or 50 plants of 60 tested). High resistance to pathotype P1 was also found in line HL08 (62.5 %, 35 of 56 tested), HL07 (60 %, 36 of 60 tested), HL05 and HL10 (both 50 %, 30 of 60 tested). Plants resistant to the pathotype P2 were found in seven hybrid lines, with the highest percent of resistant plants in HL11 (26.7 %, 16 of 60 tested). The most resistant hybrid to the pathotype P3 was line HL02, with 36.7 % resistant plants (22 of 60 tested). There were nine hybrid lines with different levels of resistance to pathotype P3. The same number of lines showed resistance to the pathotype P4, and the line with the highest number of resistant plants was again HL06 (25 %, 15 plants of 60). This line is regarded as very promising as it had a nearly fixed resistance to the pathotype P1, as reported above. There were four lines bearing some level of resistance to the pathotype P5, with HL04, where 8 plants showed stunting of roots and 7 plants showed a few minute galls on roots, which was also regarded as a resistant reaction.
In the F5 generation resistance to pathotype P1 has been fixed in HL05, HL06 and HL07 hybrid lines (Table 3). Lines HL06 and HL07 were also more resistant to P4 and P2 respectively. Higher levels of resistance to P2 has been also found in hybrid line HL10. In lines HL08 and HL13 the resistance to P3 has been greatly increased, although it was still lower compared to B. napus cv. ‘Mendel’. In contrast to cv. ‘Mendel’, none of the lines was resistant to P5.
There was a high correlation between the results of parental forms as well as susceptible and resistant standards, the Pearson’s correlation coefficient was 0,865 and it was significant at α ≤ 0.05. When calculated separately, the correlation coefficient for parental lines B. rapa ssp. pekinensis and B. napus ‘MS8’ was 0.725 and 0.765 respectively, whereas for the standards of susceptibility and resistance it was 0.980 and 0.990.
In 10 out of 30 cases (33 %) the resistance to clubroot in F4 generation was significantly higher as compared to the results of the assessment of F5 generation. The increase of resistance to the pathotype P1 was obtained in HL05, HL07 and HL10 hybrid lines and in HL08 the decrease of resistance was also significant (Fig. 3). In case of F4 and F5 generations the significant increase in disease resistance to the pathotype P2 was achieved in lines HL07 and HL10 and the other four lines remained the same (Fig. 3). Regarding the pathotypes P3 and P4 there were both times two hybrid lines with higher resistance (HL08 and HL13 as well as HL06 and HL10, respectively) and both times it was one line with decreased resistance (HL10 and HL13). Statistical analysis conducted with Kruskal – Wallis test are presented in supplementary data (Tables ST 1- ST 10).
Comparison of the resistance of standard, parental and selected hybrid genotypes to five pathotypes of Plasmodiophora brassicae (P1-P5) in F4 and F5 generations (*statistically significant differences between F4 and F5 generation of each genotype at p < 0.05; BRG – B. rapa ssp. c hinensis ‘Granaat’; MS8- male sterile line of B. napus F8 generation; HL – hybrid line; BRP – B. rapa ssp. p ekinensis; MEN – B. napus cultivar ‘Mendel’)
Seed quality
The oil content in seeds of the parental forms was 33.55 % in B. rapa ssp. pekinensis and 39.74 % in B. napus ‘MS8’. In four hybrid lines, i.e. HL10-HL14, the oil content in seeds was lower than in B. napus. In six hybrid lines HL02-HL03 and HL05-HL08, this parameter was higher than in B. rapa ssp. pekinensis. In none of the lines did the oil content exceed that of B. napus ‘MS8’ (Table 4). The protein content in seeds of the parental forms was 18.41 % in B. rapa ssp. pekinensis, and 20.13 % in B. napus ‘MS8’. Although all studied HL lines had protein content higher than both parents, only in lines HL03, HL12 and HL13 was this parameter significantly higher than in the parental forms (Table 4). The Pearson’s correlation coefficient between oil and protein content in all lines (including the two parental genotypes) was −0.439, whereas in HL lines the negative correlation between these two main parameters of seed quality was even stronger (−0.501).
The mean sinapine content in all analyzed hybrids lines was intermediate between parental genotypes. In parental forms, the sinapine content was as low as 0.91 % in B. rapa ssp. pekinensis and as high as 2.04 % in B. napus ‘MS8’ line, while in hybrid lines it ranged from 1.30 to 1.52 % (Table 4).
Use of known chromosome markers to identify brassica chromosomes
Cytogenetic analysis was carried out on the diploid plant B. rapa ssp. pekinensis (A-genome; a paternal form) and three synthetic allotetraploid plants: B. napus ‘MS8’ (AC-genome; a maternal form), high oil and protein hybrid line HL06 (AC-genome) and low oil hybrid line HL14 (AC-genome), two individuals of the F4 generation (Fig. 4 and Table 5). Investigation of mitotic chromosomes showed that the somatic complement of B. rapa spp. pekinensis had eight 5S rDNA loci and 10 35S rDNA loci (Fig. 4a). Due to co-localization of rDNA loci in some chromosomes, the two rDNA probes provided landmarks for a total of 10 A-genome chromosomes. The A3 chromosome contains the nucleolus organizer region (NOR) and usually has a distended secondary constriction in its short arm. It has two adjacent sites of 5S rDNA and 35S rDNA in its NOR. Two pairs of A1 chromosomes also have the two kinds of ribosomal RNA genes closely linked and located proximal to the centromere. A single pericentromeric locus of 35S rDNA is characteristic for the two pairs of chromosomes type VIII (cytogenetically indistinguishable A5, A6 and A9 chromosomes), while a short arm terminal locus of 5S rDNA is typical for the pair of A10 chromosomes (Fig. 4a). The 18 chromosomes of B. oleracea are shown in Fig. 4b. One pair bears the secondary constriction with a large 35S rDNA locus in the short arm of C8 chromosome. Another pair of a similar morphology, the C7 chromosome, also has a locus in the short arm, but the signal is usually less pronounced than in the C8 chromosome. One pair of C4 chromosomes exhibits a 5S rDNA signal in the proximal region of the long arm and these are two adjacent bands (Fig. 4b). In B. napus ‘MS8’, we observed 12 35S rDNA signals (Fig. 4c).
FISH analyses of somatic metaphase chromosomes of diploid B. rapa ssp. p ekinensis (a), B. oleracea (b), and synthetic allotetraploids derived from B. rapa ssp. c hinensis × B. oleracea var. gemmifera hybrid (‘MS8’ line; c-d) and B. napus ‘MS8’ × B. rapa ssp. p ekinensis hybrid (HL06; e-f and HL14; g-h). rDNA-FISH images (a, b, c, e, g) were created using probes as follows: (i) 5S rDNA labelled with rhodamine (red) and (ii) 26S rDNA labelled with digoxigenin and detected by anti-digoxigenin conjugated with FITC (green). BAC-FISH images (d, f, h) were created using BoB014O06 C-genome-specific probe labelled with digoxigenin and detected by anti-digoxigenin conjugated with FITC (green); chromosomes after rDNA-FISH and BAC-FISH were counterstained with DAPI (blue). FISH images (e-f) are marked by white arrows indicating A7/C6 recombinant chromosome and by the white lines with intervals indicating recombination breakpoints. The nomenclature of rDNA-bearing chromosomes (Arabic numerals) follows the system of the Multinational Brassica Genome Project (MBGP) Steering Committee Meeting, while linkage group assignments of the A- and C-genome chromosomes were done using A1-A10 and C1-C9, respectively (http://www.brassica.info/information/lg_assigments.htm). The Roman numerals (VIII) represent cytogenetically indistinguishable chromosomes (A5, A6 and A9). Uppercase letters denote the genomic origin of tagged chromosomes. Scale bars represent 5 μm
Hybridization with the BoB014O06 BAC probe revealed that 8 signals of 35S rDNA were located on A-genome-like and 4 other ones on C-genome-like chromosomes (Fig. 4d).
Ten 5S rDNA sites were counted with eight loci on A-genome-like chromosomes and two on C-genome-like ones. The origin of the latter is evidenced by staining of the C4 chromosome by BoB014O06 with dispersed signals along the chromosome, and by the 5S rDNA probe providing a highly condensed signal (similarly, two adjacent bands) close to the centromere. In the genomes of HL06 (36 chromosomes) and HL14 (38 chromosomes) individuals, 12 35S rDNA and 10 5S rDNA sites were detected, which is the expected rDNA loci pattern (Fig. 4e and g). To determine the parental origin of rDNA loci in hybrids, we also used a B. oleracea BAC-based probe (BoB014O06), which hybridized to all B. oleracea chromosomes showing dispersed green signals, including the ones carrying 35S rDNA (C7, C8) and 5S rDNA loci (C4) (Fig. 4e-h). However, closer inspection of the chromosomes using the BoB014O06 BAC-based probe revealed intergenomic imbalances in HL06, and we observed 19 chromosomes in the A-genome and 17 chromosomes in the C-genome (16 complete and one recombined). One non-rDNA A-genome-like chromosome was stained with BoB014O06, indicating the presence of an intergenomic chromosomal translocation, probably involving the A7 chromosome (one of the smallest A-genome chromosomes). Unlike the previous B. napus genotype, the species-specific BAC probe hybridized to the 18 C-genome-like chromosomes in the second B. napus genotype (hybrid line HL14; Fig. 4h), and there was no signal on B. rapa-like chromosomes in this individual (Fig. 4g and h). In the synthetic genome of B. napus ‘MS8’, genotype HL06 and genotype HL14, for the 35S rDNA locus, only two sites of the chromosome type VIII were found instead of the expected four sites, indicating a reduction of rDNA loci at the A-genome-like 35S rDNA, when compared with the A-genome in B. rapa spp. pekinensis.
Discussion
In the present study, 15 hybrid lines of the F4 generation were evaluated for their resistance to clubroot and plant morphology, and 10 lines showing the highest level of resistance were also tested for seed quality. The behavior of different A- and C-genome chromosomes in hybrid and parental lines was observed using advanced cytogenetic tools. Afterwards the resistant plants of 6 hybrid lines of F4 generation were pollinated under the same cover, what led to increase of plant resistance, primarily to the pathotype P1 of P. brassicae in the F5 generation.
For breeding new crops, i.e., resistant cultivars or improvement of existing cultivars, it is necessary to have diverse germplasm sources. In order to broaden the genetic base of crop species, different approaches could be employed. Species of Brassicaceae are especially amenable to genetic manipulation to improve oil type or to transfer other desired characters (Scarth and Tang 2006). Characterization is the first step was using available germplasm resources (McFerson 1998). In this study, the maternal male sterile B. napus line ‘MS8’ which was used to obtain hybrids with B. rapa ssp. pekinensis, showed heterogeneous resistance to the pathotypes P2 and P4 of P. brassicae, but it was susceptible to the pathotypes P1, P3 and P5 . The paternal B. rapa ssp. pekinensis genotype was completely resistant to pathotype P1 and it was heterogenous in its resistance to P2, P3, P4 and P5. Hybrid lines of the F4 generation, resulting from the cross between the parental lines, greatly differed in their resistance to particular pathotypes of P. brassicae. In the case of all five tested pathotypes (P1-P5) of P. brassicae, it was possible to select numerous or at least some resistant hybrid plants. Generally, the highest resistance was observed against pathotype P1, with as much as 80 % of the obtained lines (12 out of 15) being resistant. In case of the pathotypes P3-P4, both times there were nine hybrids lines showing a resistant reaction. Moreover, there were seven hybrid lines with different levels of resistance to the pathotype P2 and four hybrid lines resistant to the pathotype P5.Two hybrid lines of F4 generation, i.e. HL10 and HL14, produced plants resistant to four of five pathotypes (P1-P4). Line HL06 was resistant to just two pathotypes, but the percentage of resistant plants was very high; 83.3 % of tested plants were resistant to the pathotype P1 and 25 % of plants were resistant to the pathotype P2.
When found, it was relatively easy to fix the resistance to P1 and five such hybrid lines have been obtained, representing the F5 generation, with two resistant at levels similar to the resistant control B. napus cv. ‘Mendel’. The resistance to P1 was not combined with P3 and one may speculate that these two sources of resistance are not linked. Three lines resistant to pathotype P3 were also not associated with increased resistance to P1, which further supports this hypothesis. All in all, the resistance has been greatly improved to all pathotypes with the exception of P5, because both parental lines lacked resistance to this pathotype.
Host resistance should be used rationally to avoid development of new virulent pathotypes of the disease causal agent (Korbas et al. 2009). In clubroot, the stewardship of resistance genes is complicated by a lack of knowledge on the nature of, and relationship among, sources of resistance in commercial hybrids. In Canada, a concerted effort to produce clubroot resistant canola hybrids, led by various private companies and public breeders, has resulted in the recent release of six cultivars into the Canadian market (Pioneer ‘45H29’ and ‘D3152’, Dekalb ‘73-67RR’ and ‘73-77RR’ and Canterra ‘1960’ and Proven ‘9558C’) (Strelkov et al. 2011). In Poland, a new clubroot resistant oilseed rape variety, SY Alister has been available for at least two years (Syngenta), but the sources of resistance are not publicly available.
Surveys of clubroot resistant canola crops in Alberta, Canada, in 2013 revealed several fields in which disease incidence and severity were higher than expected for a resistant crop (Strelkov et al. 2016). Surveys of P. brassicae were made from these fields and tested for virulence on a suite of cultivars representing the various resistant canola products available in Canada. Collections of the pathogen from at least one of the fields were highly virulent on all clubroot ‘resistant’ cultivars tested, indicating that resistance was no longer effective against those strains of the pathogen.
Reports in the literature on the performance of higher generations of B. napus canola hybrids with respect to seed quality traits or their potential for heterosis in canola and rapeseed are relatively rare. In this paper, the fourth generation of hybrid lines obtained from the sexual cross B. napus (MS8) × B. rapa ssp. pekinensis was studied and found to be morphologically uniform, with the plants most closely resembling oilseed rape. In contrast, the first generation of these hybrids was intermediate, which is consistent with common observations (Mohanty et al. 2009). Grant and Beversdorf (1985) reported that high parent heterosis for oil concentration did not occur in their canola hybrids, while Sernyk and Stefansson (1983) observed that their canola hybrids performed well, but did not exceed the oil concentration of the parents. Similar results to those of Sernyk and Stefansson (1983) were obtained in this study. There were visible differences in oil content between tested hybrid lines and parents and among hybrid lines, so contrast lines with high and low rates could be selected. For instance, one tested hybrid line had only 28.32 % seed oil content, whereas the more common value for Brassicacea is usually around 30–45 % depending on the species, the variety and climatic conditions under which it is grown (El-Beltagi and Mohamed 2010). In contrast, Sabaghnia et al. (2010) reported a high degree of parent heterosis for oil concentration in canola hybrids.
In the current study, the hybrid lines displayed parent heterosis for seed protein concentration. Screening of 10 hybrid lines revealed that the protein content in hybrid lines ranged from 20.58 % to 25.15 %, while parental forms had a protein content of 18.41 % and 20.13 %. Sernyk and Stefansson (1983) found that seed oil concentration and seed protein concentration in canola were strongly negatively correlated.
Sinapine is an anti-nutritional component affecting the quality of canola meal and it has several undesirable properties as a constituent in animal feeds. It is a bitter tasting compound, making it less palatable to animals, while its presence in the diet of certain brown egg laying hens at levels exceeding 1 g/kg leads to a fishy odour or taste in the eggs (Goh et al. 1985). The elimination of sinapine content will improve the flavor, palatability and nutritional properties of canola seeds and canola meal. Although several methods for removing sinapine have been reported (Wojciechowski et al. 1994), none has proven economical thus far (Wang et al. 1998). Conventional plant breeding or genetic engineering would be a more efficient long term means of lowering or eliminating sinapine levels. The success of a conventional plant breeding program depends on finding genetic variability for sinapine content and developing a suitable analytical methodology to select plants with low sinapine content in the seed. Average sinapine concentrations among the tested hybrid lines ranged from 1.30 % to 1.52 %. There were no significant differences in sinapine content between hybrid lines, but they had significantly lower sinapine content in seeds than their maternal forms. Our results were similar to those reported in earlier studies by Niemann et al. (2012).
The most crucial aspect in the selection of lines is to ensure as good stacking of desirable characters as possible. In the current study, there were three hybrid lines combining very high levels of resistance with appropriate plant morphology and good seed quality traits. Hybrid lines HL06 and HL07 combined high resistance to pathotype P1 with a high oil content and satisfactory amount of proteins as well as low sinapine content in seeds. The hybrid line HL03 was resistant to the pathotypes P1 and P4 and also contained a relatively high amount of oil and high content of proteins and a low amount of sinapine. Such plant materials are very good sources for further improvement, by crossing with high yielding and high oil content breeding materials, provided the plants have stable numbers of chromosomes.
Amphidiploid rapeseed is widely cultivated as an important oilseed crop in many countries worldwide. Searching for forms with improved traits is highly desirable and from that point of view, interspecific crossing is a valuable tool for widening the variability of useful traits, e.g., seed quality and resistance to diseases such as clubroot, which is known as damaging to oilseed rape and vegetable brassicas (Dixon 2009a). The main sources of resistance used to date originate from different species of the genus Brassica, including B. rapa (A-genome), B. oleracea (C-genome) and B. napus (AC-genome). Different experimental approaches have been applied to study chromosome rearrangements in the Brassica allotetraploid and ancestral genomes, such as the production of synthetic allopolyploids relative to natural forms, using chromosome mapping and cytogenetic analysis including FISH (Leflon et al. 2006; Książczyk et al. 2011; Xiong et al. 2011; Fredua-Agyeman et al. 2014; Grandont et al. 2014). Physical mapping of 5S and 18S–5.8S–26S (35S) rRNA genes by FISH provides valuable chromosomal landmarks, and their characteristic positions enable chromosome identification allowing detection of chromosome variability (Maluszynska and Heslop-Harrison 1993; Hasterok et al. 2006). It also revealed a high degree of polymorphism in A-genome-like rDNA loci in successive generations of the B. napus × B. rapa ssp. pekinensis hybrids with known resistance to clubroot.
The presence of intergenomic chromosome translocations in the hybrid genome, indicating that the two parental genomes may have undergone some rearrangements following hybridization, was revealed in newly synthesized Brassica allopolyploids (Książczyk et al. 2011; Xiong et al. 2011), which can be a rapid response to formation of the allotetraploid genome. The use of a species-specific B. oleracea BAC clone revealed the chromosome rearrangements between A- and C-genomes in the synthetic B. napus forms (Książczyk et al. 2011), and our present work confirmed this observation presenting possible A7/C6 chromosome translocation in the B. napus genotype HL06. Similarly, an A7/C6 chromosomal translocation was observed in synthetic Brassica allotetraploids by Xiong et al. (2011) and Grandont et al. (2014), indicating known patterns of genome duplication within the Brassica napus genome (Parkin et al. 2003). It is worth mentioning that none of the rDNA-bearing chromosomes were involved in recombination showing the A7/C6 translocation, because the number of rDNA loci is stable and A-genome-like chromosomes are not painted by a C-genome-specific BAC clone. Other A/C recombinations changing the rDNA loci pattern, showing locus gain or loss, would also be possible.
The base chromosomal types have been introduced and described in detail by Hasterok et al. (2001). These types were used originally to describe the rDNA loci patterns in the three genomes (A, B and C) that constitute the six most studied crop species of Brassica. Comparative analyses revealed that variation in the number and chromosomal position of 5S and 35S rDNA occurs and that only chromosomes with proximally and/or pericentromerically distributed ribosomal rRNA genes were polymorphic (Hasterok et al. 2006). These chromosomes represented types II and VIII and it seems that the same chromosome may occur in different materials as type II or type VIII, as they were observed in three of six B. rapa accessions (Hasterok et al. 2006), leading to the more frequent occurrence of type VIII compared with type II chromosomes. In the present work, we observed two instead of the expected four 35S rDNA sites pericentromerically located in B. napus chromosomes, indicating a reduction in rDNA loci carried by the A genome corresponding to the chromosomal type VIII. It can be concluded that more detailed FISH analyses of mitotic chromosomes and their rearrangements will be required beyond study of ribosomal landmarks. This could be accomplished through the use of either chromosome-specific or even arm-specific sets of BAC clone-based probes for both the B. rapa and B. oleracea chromosomes, together with PCR-based disease resistance markers (Koo et al. 2004; Xiong et al. 2011; Fredua-Agyeman et al. 2014).
References
Agrios, G. N. (2005). Plant Pathology 5th edition (pp. 922). San Diego: Elsevier-Academic Press.
Allard, R. W. (1960). Principles of plant breeding. New York: Wiley & Sons Inc..
Bagget, J. R., & Kean, D. (1985). Clubroot resistant broccoli breeding lines OSU CR-2 to OSU CR-8. Horticulturae Science, 20, 784.
Diederichsen, E., & Sacristán, M. D. (1996). Disease response of resynthesized Brassica napus L. Lines carrying different combinations of resistance to Plasmodiophora brassicae Wor. Plant Breeding, 115, 5–10.
Diederichsen, E., Beckmann, J., Schondelmeier, J., & Dreyer, F. (2006). Genetics of clubroot resistance in Brassica napus ‘Mendel’. Acta Horticulturae, 706, 307–311.
Diederichsen, E., Frauen, M., Linders, E., Hatakeyama, K., & Hirai, M. (2009). Status and perspectives of clubroot resistance breeding in crucifer crops. Journal of Plant Growth Regulation, 28, 265–281.
Dixon, G. R. (2009a). The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. Journal of Plant Growth Regulation, 28, 194–202.
Dixon, G. R. (2009b). Plasmodiophora brassicae In its environment. Journal of Plant Growth Regulation, 28, 212–228.
El-Beltagi, H. E. S., & Mohamed, A. A. (2010). Variations in fatty acid composition, glucosinolate profile and some phytochemical contents in selected oil seed rape (Brassica napus L.) cultivars. Grasas y Aceites, 61, 143–150.
Engqvist, L. G. (1994). Distribution of clubroot (Plasmodiophora brassicae Wor.) in Sweden and the effect of infection on oil content of oilseed rape (Brassica napus L.). Journal of the Swedish Seed Association, 104, 82–86.
European Union. (2009). Common Catalogue of Varieties of Agricultural Plant Species. 28th Complete Edition (2009/C 302 A/01), Official Journal of the European Union 12.12.2009.
Fredua-Agyeman, R., Coriton, O., Huteau, V., Parkin, I. A. P., Chèvre, A. M., & Rahman, H. (2014). Molecular cytogenetic identification of B genome chromosomes linked to blackleg disease resistance in Brassica napus × B. carinata interspecific hybrids. Theoretical and Applied Genetics, 127, 1305–1318.
Gerlach, W. L., & Dyer, T. A. (1980). Sequence organisation of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Research, 8, 4851–4865.
Goh, Y.K., Robblee, A.R., & Clandinin, D.R. (1985). Influence of glucosinolates and free oxazolidinethione in a laying diet containing a constant amount of sinapine on the thyroid size and hepatic trimethylamine oxidase activity of brown-egg layers. Canadian Journal of Animal Sciences, 65, 921─927.
Grandont, L., Cuňado, N., Coriton, O., Huteau, V., Eber, F., Chèvre, A. M., Grelon, M., Chelysheva, L., & Jenczewski, E. (2014). Homoeologous chromosome sorting and progression of meiotic recombination in Brassica napus: ploidy does matter! Plant Cell. doi:10.1105/tpc.114.122788.
Grant, I., & Beversdorf, W. D. (1985). Heterosis and combining ability estimates in spring-planted oilseed rape (Brassica napus L.). Canadian Journal of Genetics and Cytology, 27, 472–478.
Hasterok, R., Jenkins, G., Langdon, T., Jones, R. N., & Maluszynska, J. (2001). Ribosomal DNA is an effective marker of Brassica chromosomes. Theoretical and Applied Genetics, 103, 486–490.
Hasterok, R., Wolny, E., Hosiawa, M., Kowalczyk, M., Kulak-Książczyk, S., Książczyk, T., Heneen, W. K., & Maluszynska, J. (2006). Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Annals of Botany, 97, 205–216.
Hirai, M. (2006). Genetic analysis of clubroot resistance in Brassica rapa. Breeding Science, 56, 223–229.
Howell, E. C., Barker, G. C., Jones, G. H., Kearsey, M. J., King, G. J., Kop, E. P., Ryder, C. D., Teakle, G. R., Vicente, J. G., & Armstrong, S. J. (2002). Integration of the cytogenetic and genetic linkage maps of Brassica oleracea. Genetics, 161, 1225–1234.
Jedryczka, M., Korbas, M., Jajor, E., Danielewicz, J., & Kaczmarek, J. (2013). The occurrence of Plasmodiophora brassicae in agricultural soils in Wielkopolska region, in 2011–2012 [in polish with English abstract]. Progress in Plant Protection/Postępy w Ochronie Roślin, 53, 774–778.
Jedryczka, M., Kasprzyk, I., Korbas, M., Jajor, E., & Kaczmarek, J. (2014). Infestation of polish agricultural soils by Plasmodiophora brassicae along the polish-Ukrainian border. Journal of Plant Protection Research, 54, 238–241.
Kaczmarek, J., Latunde-Dada, A. O., Irzykowski, W., Cools, H. J., Stonard, J. F., Brachaczek, A., & Jedryczka, M. (2014). Molecular screening for avirulence alleles AvrLm1 and AvrLm6 in airborne inoculum of Leptosphaeria maculans and winter oilseed rape (Brassica napus) plants from Poland and the UK. Journal of Applied Genetics, 5, 529–539.
Kim, H., Choi, S. R., Bae, J., Hong, C. P., Lee, S. Y., Hossain, M. J., Van Nguyen, D., Jin, M., Park, B. S., Bang, J. W., Bancroft, I., & Lim, Y. P. (2009). Sequenced BAC anchored reference genetic map that reconciles the ten individual chromosomes of Brassica rapa. BMC Genomics, 10, 432. doi:10.1186/1471-2164-10-432.
Konieczny, W. (2012). Clubroot is present on 250 thousand hectares [in polish]. Farmer, 5, 38–42.
Koo, D. H., Plaha, P., Lim, Y. P., Hur, Y., & Bang, J. W. (2004). A high resolution karyotype of Brassica rapa ssp. pekinensis revealed by pachytene analysis and multicolour fluorescence in situ hybridisation. Theoretical and Applied Genetics, 109, 1346–1352.
Korbas, M., Jajor, E., & Budka, A. (2009). Clubroot (Plasmodiophora brassicae) – a threat for oilseed rape. Journal of Plant Protection Research, 49, 446–451.
Korbas, M., Jajor, E., Kaczmarek, J., Perek, A., & Jedryczka, M. (2014). Infestation of polish agricultural soils by Plasmodiophora brassicae on the polish-Belarussian border in Podlasie province. Integrated Control in Oilseed Crops. IOBC/wprs Bulletin, 104, 167–171.
Kruskal, W. H. (1952). A nonparametric test for the several sample problem. The Annals of Mathematical Statistics, 23, 525–540.
Książczyk, T., Taciak, M., & Zwierzykowski, Z. (2010). Variability of ribosomal DNA sites in Festuca pratensis, Lolium perenne, and their intergeneric hybrids, revealed by FISH and GISH. Journal of Applied Genetics, 51, 449–460.
Książczyk, T., Kovarik, A., Eber, F., Huteau, V., Khaitova, L., Tesarikova, Z., Coriton, O., & Chèvre, A. M. (2011). Immediate unidirectional epigenetic reprogramming of NORs occurs independently of rDNA rearrangements in synthetic and natural form of a polyploid species Brassica napus. Chromosoma, 120, 557–571.
Kuginuki, Y., Yoshikawa, H., & Hirai, M. (1999). Variation in virulence of Plasmodiophora brassicae in Japan tested with clubroot-resistant cultivars of Chinese cabbage (Brassica rapa L. Ssp. pekinensis). European Journal of Plant Pathology, 105, 327–332.
Kutcher, R., Balesdent, M. H., Rimmer, S. R., Rouxel, T., Chevre, A. M., Delourme, R., & Brun, H. (2010). Frequency of avirulence genes in Leptosphaeria maculans in western Canada. Canadian Journal of Plant Pathology, 32, 77–85.
Leflon, M., Eber, F., Letanneur, J. C., Chelysheva, L., Coriton, O., Huteau, V., Ryder, C. D., Barker, G., Jenczewski, E., & Chevre, A. M. (2006). Pairing and recombination at meiosis of Brassica rapa (AA) × Brassica napus (AACC) hybrids. Theoretical and Applied Genetics, 113, 1467–1480.
Li, H., Sivasithamparam, K., & Barbetti, M. J. (2003). Breakdown of a B. rapa ssp. sylvestris single dominant resistance gene in B. napus by L. maculans field isolates. Plant Disease, 87, 752.
Lüders, W., Abel, S., Friedt, W., Kopahnke, D., & Ordon, F. (2011). Auftreten von Plasmodiophora brassicae als Erreger der Kohlhernie im Winterrapsanbau in Europa sowie Identifizierung, Charakterisierung und molekulare Kartierung neuer Kohlhernieresistenzgene aus genetischen Ressourcen Drittes Nachwuchswissenschaftler-forum, 23–25 November 2010, Quedlinburg. Julius-Kühn-Archiv., 430, 40–43.
Maluszynska, J., & Heslop-Harrison, J. S. (1993). Physical mapping of rDNA loci in Brassica species. Genome, 36, 774–781.
McFerson, J. R. (1998). From in situ to ex situ and back: the importance of characterizing germplasm collections. Hortscience, 33, 1134–1135.
Mohanty, A., Chrungu, B., Verma, N., & Shivanna, K. R. (2009). Broadening the Genetic Base of crop brassicas by production of new intergeneric hybrid. Czech Journal of Genetics and Plant Breeding, 45, 117–122.
Moreno-Gonzalez, J., & Cubero, J. I. (1993). Selection strategies and choice of breeding methods. London: Chapman&Hall.
Niemann, J., Wojciechowski, A., & Janowicz, J. (2012). Broadening the variability of quality traits in rapeseed through interspecific hybridization with an application of immature embryo culture. Journal of Biotechnology, Computational Biology and Bionanotechnology, 3, 109–115.
Nomura, K., Minegishi, Y., Kimizuka-Takagi, C., Fujioka, T., Moriguchi, K., Shishido, R., & Ikehashi, H. (2005). Evaluation of F2 and F3 plants introgressed with QTL for clubroot resistance in cabbage developed by using SCAR markers. Plant Breeding, 124, 371–375.
Pageau, D., Lajeunesse, J., & Lafond, J. (2006). Impact de l’hernie des crucife’res [Plasmodiophora brassicae] Sur la productivite `et la qualite `du canola. Canadian Journal of Plant Pathology, 28, 137–143.
Parkin, I. A. P., Sharpe, A. G., & Lydiate, D. J. (2003). Patterns of genome duplication within the Brassica napus genome. Genome, 146, 291–303.
Piao, Z., Ramchiary, N., & Lim, Y. P. (2009). Genetics of clubroot resistance in Brassica species. Journal of Plant Growth Regulation, 28, 252–264.
Rahman, H., Peng, G., Yu, F., Falk, K. C., Kulkarni, M., & Selvaraj, G. (2014). Genetics and breeding for clubroot resistance in Canadian spring canola (Brassica napus L.). Canadian Journal of Plant Pathology, 36, 122–134.
Ričařová, V., Kaczmarek, J., Strelkov, S. E., Kazda, J., Lueders, W., Rysanek, P., Manolii, V., & Jedryczka, M. (2016). Pathotypes of Plasmodiophora brassicae causing damage to oilseed rape in the Czech Republic and Poland. European Journal of Plant Pathology, 145, 559–572.
Robak, J. (1991). Variability of the pathotypes of Plasmodiophora brassicae Wor, Present in Poland and Their Pathogenicity to the Cultivars and Breeding Lines of Brassica oleracea [in Polish]. Habilitation monograph. (pp. 59). Skierniewice: Instytut Warzywnictwa.
Robak, J. (1994). Crop rotation effect on clubroot disease decrease. Acta. Horticulturae (ISHS), 371, 223–226.
Rouxel, T., Penaud, A., Pinochet, X., Brun, H., Gout, L., Delourme, R., Schmit, J., & Balesdent, M. H. (2003). A ten-year survey of populations of Leptosphaeria maculans in France indicates a rapid adaptation towards the Rlm1 resistance gene in oilseed rape. European Journal of Plant Pathology, 109, 871–881.
Sabaghnia, N., Dehghani, H., Alizadeh, B., & Mohghaddam, M. (2010). Heterosis and combining ability analysis for oil yield and its components in rapeseed. Austalian Journal of Crop Science, 4, 390–397.
Scarth, R., & Tang, G. (2006). Modification of Brassica oil using conventional and transgenic approaches. Crop Science, 46, 1225–1236.
Sernyk, J. L., & Stefansson, B. R. (1983). Heterosis in summer rape. Canadian Journal of Plant Science, 63, 407–413.
Somé, A., Manzanares, M. J., Laurens, F., Baron, F., Thomas, G., & Rouxel, F. (1996). Variation for virulence on Brassica napus L. Amongst Plasmodiophora brassicae collections from France and derived single-spore isolates. Plant Pathology, 45, 432–439.
Stachowiak, A., Olechnowicz, J., Jędryczka, M., Rouxel, T., Balesdent, M. H., Happstadius, I., Gladders, P., Latunde-Dada, A., & Evans, N. (2006). Frequency of avirulence alleles in field populations of Leptosphaeria maculans in Europe. European Journal of Plant Pathology, 114, 67–75.
Strelkov, S. E., Hwang, S. F., Howard, R. J., & Turkington, T. K. (2011). Progress Towards the Sustainable Management of Clubroot [Plasmodiophora brassicae] of Canola on the Canadian Prairies. Insects and Diseases: Prairie Soils & Crops Journal, 4, 114–121.
Strelkov, S. E., Hwang, S. F., Manolii, V. P., Cao, T., & Feindel, D. (2016). Emergence of new virulence phenotypes of Plasmodiophora brassicae on canola (Brassica napus) in Alberta, Canada. European Journal of Plant Pathology, 145, 517–529.
Suwabe, K., Tsukazaki, H., Iketani, H., Hatakeyama, K., & Fujimura, M. (2003). Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theoretical and Applied Genetics, 107, 997–1002.
Tewari, J. P., & Mithen, R. F. (1999). Diseases. In Gomez-Campo (Ed.), Biology of Brassica coenospicies, developments in plant genetics and breeding 4 (pp. 375–413). Amsterdam: Elsevier Science B. V..
Unfried, I., & Gruendler, P. (1990). Nucleotide sequence of the 5.8S and 25S rRNA genes and the internal transcribed spacers from Arabidopsis thaliana. Nucleic Acids Research, 18, 4011.
Van de Wouw, A. P., Stonard, J. F., Howlett, B. J., West, J. S., Fitt, B. D. L., & Atkins, S. D. (2010). Determining frequencies of avirulent alleles in airborne Leptosphaeria maculans inoculum using quantitative PCR. Plant Pathology, 59, 809–818.
Voorrips, R. E. (1995). Plasmodiophora brassicae: aspects of pathogenesis and resistance in Brassica oleracea. Euphytica, 83, 139–146.
Wallenhammar, A. C., Johnsson, L., & Gerhardson, B. (1999). Clubroot resistance and yield loss in spring oilseed turnip rape and spring oilseed rape. Canberra: Proc. 10th Int. Rapeseed Congress.
Wang, S. X., Oomah, B. D., Ian McGregor, D., & Downey, R. K. (1998). Genetic and seasonal variation in the sinapine content of seed from Brassica and Sinapis species. Canadian Journal of Plant Science, 78, 395–400.
Wit, F., & Van de Weg, M. (1964). Clubroot resistance in turnips (Brassica campestris L.) I. Physiological races of the parasite and their identification in mixtures. Euphytica, 13, 9–18.
Wojciechowski, A. (1993). Some morphological and phenological traits and fertility of lines of artificial winter oilseed rape originated from male sterile plants (Brassica napus var. oleifera). Genetica Polonica, 34, 317–325.
Wojciechowski, A., Kott, L., & Beversdorf, W. (1994). Content of sinapine in haploid embryos of rapeseed (Brassica napus) from microspore in in vitro culture. Rośliny Oleiste – Oilseed Crops, 15, 105–110.
Xiong, Z., Gaeta, R. T., & Pires, J. C. (2011). Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proceedings of the National Academy of Science USA, 108, 7908–7913.
Yoshikawa, H. (1981). Breeding for clubroot resistance in Chinese cabbage. In N. S. Taleker & T. D. Griggs (Eds.), Chinese cabbage (pp. 405–413). Shanhua: AVRDC.
Acknowledgments
This project was funded by the National Research Centre N N310 298439. The authors wish to thank Plant Breeding Strzelce, Branch Malyszyn, Poland, for the seed quality analyses. We also thank Prof. S.E. Strelkov, University of Alberta, Canada and Dr. William Truman from the Institute of Plant Genetics of the Polish Academy of Sciences, for thorough language corrections.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
ESM 1
(DOC 369 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Niemann, J., Kaczmarek, J., Książczyk, T. et al. Chinese cabbage (Brassica rapa ssp. pekinensis) – a valuable source of resistance to clubroot (Plasmodiophora brassicae). Eur J Plant Pathol 147, 181–198 (2017). https://doi.org/10.1007/s10658-016-0991-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-016-0991-x