Abstract
Scan statistics have been used extensively to identify temporal clusters of health events. We describe the temporal cluster detection methodology adopted by the EUROCAT (European Surveillance of Congenital Anomalies) monitoring system. Since 2001, EUROCAT has implemented variable window width scan statistic for detecting unusual temporal aggregations of congenital anomaly cases. The scan windows are based on numbers of cases rather than being defined by time. The methodology is imbedded in the EUROCAT Central Database for annual application to centrally held registry data. The methodology was incrementally adapted to improve the utility and to address statistical issues. Simulation exercises were used to determine the power of the methodology to identify periods of raised risk (of 1–18 months). In order to operationalize the scan methodology, a number of adaptations were needed, including: estimating date of conception as unit of time; deciding the maximum length (in time) and recency of clusters of interest; reporting of multiple and overlapping significant clusters; replacing the Monte Carlo simulation with a lookup table to reduce computation time; and placing a threshold on underlying population change and estimating the false positive rate by simulation. Exploration of power found that raised risk periods lasting 1 month are unlikely to be detected except when the relative risk and case counts are high. The variable window width scan statistic is a useful tool for the surveillance of congenital anomalies. Numerous adaptations have improved the utility of the original methodology in the context of temporal cluster detection in congenital anomalies.




Similar content being viewed by others
References
Naus J, Wallenstein S. Temporal surveillance using scan statistics. Stat Med. 2006;25(2):311–24.
Dolk H, Loane M, Garne E. The prevalence of congenital anomalies in Europe. Adv Exp Med Biol. 2010;686:349–64.
Khoshnood B, Greenlees R, Loane M, Dolk H. Paper 2: EUROCAT public health indicators for congenital anomalies in Europe. Birth Defects Res A Clin Mol Teratol. 2011;91(Suppl 1):S16–22.
Boyd PA, Haeusler M, Barisic I, Loane M, Garne E, Dolk H. Paper 1: the EUROCAT network–organization and processes. Birth Defects Res A Clin Mol Teratol. 2011;91(Suppl 1):S2–15.
Loane M, Dolk H, Kelly A, Teljeur C, Greenlees R, Densem J. Paper 4: EUROCAT statistical monitoring: identification and investigation of 10 year trends of congenital anomalies in Europe. Birth Defects Res A Clin Mol Teratol. 2011;91(Suppl 1):S31–43.
Dolk H, Loane M, Teljeur C, Densem J, Greenlees R, McCullough N et al. Detection and investigation of temporal clusters of congenital anomaly in Europe: seven years of experience of the EUROCAT surveillance system. Epidemiology 2015 [Epub ahead of print, PMID: 25840712).
Centres for Disease Control and Prevention. Guidelines for investigating clusters of health events. Morbidity and mortality weekly report 1990; 39(RR-11):1–16.
Sego LH, Woodall WH, Reynolds MR. A comparison of surveillance methods for small incidence rates. Stat Med. 2008;27(8):1225–47.
Quataert PK, Armstrong B, Berghold A, Bianchi F, Kelly A, Marchi M, Martuzzi M, Rosano A. Methodological problems and the role of statistics in cluster response studies: a framework. Eur J Epidemiol. 1999;15(9):821–31.
Tango T. Tests for temporal clustering. In: Gail M, Krickeberg K, Samet J, Tsiatis A, Wong W, editors. Statistical methods for disease clustering. New York: Springer; 2010. p. 49–70.
Nagarwalla N. A scan statistic with a variable window. Stat Med. 1996;15(7–9):845–50.
Naus JI. The distribution of the size of the maximum cluster of points on a line. J Am Stat Assoc. 1965;60(310):532–8.
Glaz J, Zhang Z. Multiple window discrete scan statistics. J Appl Stat. 2004;31(8):967–80.
McCullough N, Loane M, Greenlees R, Dolk H. EUROCAT statistical monitoring report—2009. 2012. Newtown abbey, Northern Ireland, EUROCAT Central Registry.
Kulldorff M. SaTScan user guide for version 9.3. 2014. Boston, USA.
Weinstock MA. A generalised scan statistic test for the detection of clusters. Int J Epidemiol. 1981;10(3):289–93.
Unkel S, Farrington CP, Garthwaite PH, Robertson C, Andrews N. Statistical methods for the prospective detection of infectious disease outbreaks: a review. J R Stat Soc Ser A Stat Soc. 2012;175(1):49–82.
Fricker RD. Some methodological issues in biosurveillance. Stat Med. 2011;30(5):403–15.
Tango T. A test for spatial disease clustering adjusted for multiple testing. Stat Med. 2000;19(2):191–204.
Cucala L. A hypothesis-free multiple scan statistic with variable window. Biom J. 2008;50(2):299–310.
Acknowledgments
We wish to thank Joan Morris and Fabrizio Bianchi for their helpful comments in an earlier draft of this manuscript. EU Commission Public Health Programme EUROCAT Joint Action 2011–2013 (Grant No. 2010 22 04).
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix: The scan statistic defined by Nagarwalla [11]
Appendix: The scan statistic defined by Nagarwalla [11]
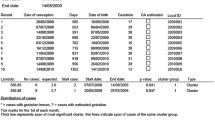
An analysis requires the dates of a set of cases, and the specification of the start and end dates for the analysis period. For a given subset of n cases occurring between date start and date end , it is possible to calculate the test statistic lambda as follows:
Where r is the total number of cases, and
The dates date first and date last are the dates of the first and last cases in the scanning window, respectively. Typically a minimum scanning window of 5 is used. Lambda is calculated for every possible subset of n consecutive cases for n = 5,…, r
The lambda for each subset of cases is calculated and the largest found is recorded along with the details of the start and end dates. Next, simulated datasets are generated each with r uniform random numbers in the interval [0,1]. For each simulation, the lambda for each subset is calculated and the largest lambda is recorded. After, for example, 999 iterations the lambdas are ordered by size from smallest to largest and the 95th percentile is noted as λsig. If the lambda from the real dataset is greater than to λsig then the cases are designated as a significant cluster.
The method assumes a uniform distribution of cases and hence no seasonal variation.
Multiple cases in the observed data can occur on the same day. Where the number of cases in a single day is equal to or greater than the minimum window size, the d value for that window will equal zero and hence lambda will equal infinity. A nominal value equivalent to an hour is added to each case that occurs on the same day, thereby distributing multiple cases. An upper limit of 24 cases within a day is a reasonable assumption in our context. The alternative of distributing same-day cases uniformly across 24 h would, in theory, be preferred.
It should be noted that different statistical terminology was adopted in EUROCAT to avoid confusion with variable names in computer coding. Hence our notation uses ‘r’ in place of ‘N’. To maintain consistency between the code and this article, we have used same terminology in the code and the paper.
Rights and permissions
About this article
Cite this article
Teljeur, C., Kelly, A., Loane, M. et al. Using scan statistics for congenital anomalies surveillance: the EUROCAT methodology. Eur J Epidemiol 30, 1165–1173 (2015). https://doi.org/10.1007/s10654-015-0044-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10654-015-0044-3