Abstract
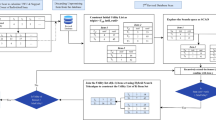
Efficient identification of cohorts of similar patients is a major precondition for personalized medicine. In order to train prediction models on a given medical data set, similarities have to be calculated for every pair of patients—which results in a roughly quadratic data blowup. In this paper we discuss the topic of in-database patient similarity analysis ranging from data extraction to implementing and optimizing the similarity calculations in SQL. In particular, we introduce the notion of chunking that uniformly distributes the workload among the individual similarity calculations. Our benchmark comprises the application of one similarity measures (Cosine similariy) and one distance metric (Euclidean distance) on two real-world data sets; it compares the performance of a column store (MonetDB) and a row store (PostgreSQL) with two external data mining tools (ELKI and Apache Mahout).













Similar content being viewed by others
References
Anthony Celi, L., Mark, R.G., Stone, D.J., Montgomery, R.A.: “Big data” in the intensive care unit. Closing the data loop. Am. J. Respir. Crit. Care Med. 187(11), 1157–1160 (2013)
Apache Mahout Committers: Apache Mahout. https://mahout.apache.org
Brown, S.A.: Patient similarity: emerging concepts in systems and precision medicine. Front. Physiol. 7, 561 (2016)
Cabrera, W., Ordonez, C.: Scalable parallel graph algorithms with matrix-vector multiplication evaluated with queries. Distrib. Parallel Databases 35(3–4), 335–362 (2017)
Chaudhuri, S., Dayal, U.: An overview of data warehousing and olap technology. ACM Sigmod Rec 26(1), 65–74 (1997)
Deza, M.M., Deza, E.: Encyclopedia of Distances. Springer, Berlin (2012)
Dheeru, D., Karra Taniskidou, E.: UCI machine learning repository (2017). http://archive.ics.uci.edu/ml
Domínguez-Muñoz, J.E., Carballo, F., Garcia, M.J., de Diego, J.M., Campos, R., Yangúela, J., de la Morena, J.: Evaluation of the clinical usefulness of apache II and saps systems in the initial prognostic classification of acute pancreatitis: a multicenter study. Pancreas 8(6), 682–686 (1993)
Drost, H.G.: R philentropy package. https://cran.r-project.org/web/packages/philentropy/philentropy.pdf
ELKI Development Team: ELKI: Environment for Developing KDD-Applications Supported by Index-Structures. https://elki-project.github.io/
Ferreira, F.L., Bota, D.P., Bross, A., Mélot, C., Vincent, J.L.: Serial evaluation of the SOFA score to predict outcome in critically ill patients. JAMA 286(14), 1754–1758 (2001)
Garcelon, N., Neuraz, A., Benoit, V., Salomon, R., Kracker, S., Suarez, F., Bahi-Buisson, N., Hadj-Rabia, S., Fischer, A., Munnich, A.: Finding patients using similarity measures in a rare diseases-oriented clinical data warehouse: Dr. Warehouse and the needle in the needle stack. J. Biomed. Inform. 73, 51–61 (2017)
Gottlieb, A., Stein, G.Y., Ruppin, E., Altman, R.B., Sharan, R.: A method for inferring medical diagnoses from patient similarities. BMC Med. 11(1), 194 (2013)
Hill, M.D., Marty, M.R.: Amdahl’s law in the multicore era. IEEE Comput. 41(7), 33–38 (2008)
Hoogendoorn, M., El Hassouni, A., Mok, K., Ghassemi, M., Szolovits, P.: Prediction using patient comparison vs. modeling: a case study for mortality prediction. In: 2016 IEEE 38th Annual International Conference of the Engineering in Medicine and Biology Society (EMBC), pp. 2464–2467 (2016)
Johnson, A.E., Pollard, T.J., Shen, L., Lehman, L.W.H., Feng, M., Ghassemi, M., Moody, B., Szolovits, P., Celi, L.A., Mark, R.G.: MIMIC-III, a freely accessible critical care database. Sci. Data 3, 160035 (2016)
Le Gall, J.R., Lemeshow, S., Saulnier, F.: A new simplified acute physiology score (SAPS II) based on a european/north american multicenter study. JAMA 270(24), 2957–2963 (1993)
Lee, J., Maslove, D.M., Dubin, J.A.: Personalized mortality prediction driven by electronic medical data and a patient similarity metric. PLoS ONE 10(5), e0127428 (2015)
Li, L., Cheng, W.Y., Glicksberg, B.S., Gottesman, O., Tamler, R., Chen, R., Bottinger, E.P., Dudley, J.T.: Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci. Transl. Med. 7(311), 311ra174–311ra174 (2015)
Morid, M.A., Sheng, O.R.L., Abdelrahman, S.: PPMF: a patient-based predictive modeling framework for early ICU mortality prediction (2017). arXiv preprint. arXiv:1704.07499
Ordonez, C.: Statistical model computation with UDFS. IEEE Trans. Knowl. Data Eng. 22(12), 1752–1765 (2010)
Ordonez, C., Cabrera, W., Gurram, A.: Comparing columnar, row and array DBMSS to process recursive queries on graphs. Inf. Syst. 63, 66–79 (2017)
Park, Y.J., Kim, B.C., Chun, S.H.: New knowledge extraction technique using probability for case-based reasoning: application to medical diagnosis. Expert Syst. 23(1), 2–20 (2006)
Passing, L., Then, M., Hubig, N., Lang, H., Michael, S., Günnemann, S., Kemper, A., Neumann, T.: SQL- and operator-centric data analytics in relational main-memory databases. In: EDBT, pp. 84–95 (2017)
Qin, C., Rusu, F.: Dot-product join: Scalable in-database linear algebra for big model analytics. In: Proceedings of the 29th International Conference on Scientific and Statistical Database Management, p. 8. ACM, New York (2017)
Raasveldt, M., Holanda, P., Mühleisen, H., Manegold, S.: Deep integration of machine learning into column stores. In: EDBT, pp. 473–476. OpenProceedings.org (2018)
Saeed, M., Villarroel, M., Reisner, A.T., Clifford, G., Lehman, L.W., Moody, G., Heldt, T., Kyaw, T.H., Moody, B., Mark, R.G.: Multiparameter intelligent monitoring in intensive care II (MIMIC-II): a public-access intensive care unit database. Crit. Care Med. 39(5), 952 (2011)
Schubert, E., Koos, A., Emrich, T., Züfle, A., Schmid, K.A., Zimek, A.: A framework for clustering uncertain data. Proc. VLDB Endow. 8(12), 1976–1979 (2015)
Sharafoddini, A., Dubin, J.A., Lee, J.: Patient similarity in prediction models based on health data: a scoping review. JMIR Med. Inform. 5(1), e7 (2017)
Strack, B., DeShazo, J.P., Gennings, C., Olmo, J.L., Ventura, S., Cios, K.J., Clore, J.N.: Impact of HbA1c measurement on hospital readmission rates: analysis of 70,000 clinical database patient records. BioMed Res. Int. 2014, 781670 (2014)
Sun, J., Sow, D., Hu, J., Ebadollahi, S.: A system for mining temporal physiological data streams for advanced prognostic decision support. In: 2010 IEEE 10th International Conference on Data Mining (ICDM), pp. 1061–1066 (2010)
Vincent, J.L., Moreno, R., Takala, J., Willatts, S., De Mendonça, A., Bruining, H., Reinhart, C., Suter, P., Thijs, L.: The SOFA (sepsis-related organ failure assessment) score to describe organ dysfunction/failure. Intensive Care Med. 22(7), 707–710 (1996)
Wang, F., Hu, J., Sun, J.: Medical prognosis based on patient similarity and expert feedback. In: 2012 21st International Conference on Pattern Recognition (ICPR), pp. 1799–1802 (2012)
Wang, S., Li, X., Yao, L., Sheng, Q.Z., Long, G.: Learning multiple diagnosis codes for ICU patients with local disease correlation mining. ACM Trans. Knowl. Discov. Data (TKDD) 11(3), 31 (2017)
Wiese, L.: Advanced Data Management for SQL, NoSQL, Cloud and Distributed Databases. DeGruyter/Oldenbourg, Munich (2015)
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wiese, I., Sarna, N., Wiese, L. et al. Concept acquisition and improved in-database similarity analysis for medical data. Distrib Parallel Databases 37, 297–321 (2019). https://doi.org/10.1007/s10619-018-7249-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10619-018-7249-x