Abstract
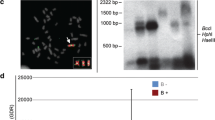
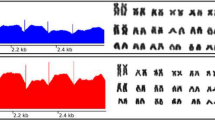
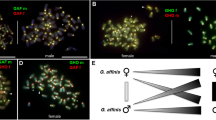
B chromosomes are dispensable elements observed in many eukaryotic species, including the African cichlid Astatotilapia latifasciata, which might have one or two B chromosomes. Although there have been many studies focused on the biology of these chromosomes, questions about the evolution, maintenance, and potential effects of these chromosomes remain. Here, we identified a variant form of the hnRNP Q-like gene inserted into the B chromosome of A. latifasciata that is characterized by a high copy number and intron-less structure. The absence of introns and presence of transposable elements with a reverse transcriptase domain flanking hnRNP Q-like sequences suggest that this gene was retroinserted into the B chromosome. RNA-Seq analysis did not show that the B variant retroinserted copies are transcriptionally active. However, RT-qPCR results showed variations in the canonical hnRNP Q-like copy expression levels among exons, tissues, sex, and B presence/absence. Although the patterns of transcription are not well understood, the exons of the B retrocopies were overexpressed, and a bias for female B+ expression was also observed. These results suggest that retroinsertion is an additional and important mechanism contributing to B chromosome formation. Furthermore, these findings indicate a bias towards female differential expression of B chromosome sequences, suggesting that B chromosomes and sex determination are somehow associated in cichlids.




Similar content being viewed by others
Abbreviations
- CDS:
-
Coding DNA sequence
- FPKM:
-
Fragments per kilobase of exon per million fragments mapped
- FR:
-
Flanking regions
- GDR:
-
Gene dose ratio
- GLM:
-
Generalized linear model
- hnRNP Q-like:
-
Heterogeneous nuclear ribonucleoprotein Q-like
- ORFs:
-
Open reading frames
- qPCR:
-
Quantitative PCR
- RIN:
-
RNA integrity number
- RT-qPCR:
-
Reverse transcription quantitative PCR
- SNVs:
-
Single nucleotide variations
- TEs:
-
Transposable elements
- UBCE:
-
Ubiquitin-conjugating enzyme
- UTRs:
-
Untranslated regions
References
Bai Y, Casola C, Feschotte C, Betran E (2007) Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in Drosophila. Genome Biol 8:R11
Banaei-Moghaddam AM, Meier K, Karimi-Ashtiyani R, Houben A (2013) Formation and expression of pseudogenes on the B chromosome of rye. Plant Cell 25:2536–2544
Banaei-Moghaddam AM, Martis MM, Macas J et al (2014) Genes on B chromosomes: old questions revisited with new tools. Biochim Biophys Acta 1849:64–70
Bel Y, Ferré J, Escriche B (2011) Quantitative real-time PCR with SYBR green detection to assess gene duplication in insects: study of gene dosage in Drosophila melanogaster (Diptera) and in Ostrinia nubilalis (Lepidoptera). BMC Res Notes 4:84
Beukeboom LW (1994) Bewildering Bs: an impression of the 1st B-chromosome conference. Heredity 73:328–336
Brawand D, Wagner CE, Li YI et al (2014) The genomic substrate for adaptive radiation in African cichlid fish. Nature 513:375–381
Burt A, Trivers R (2006) Genes in conflict: the biology of selfish genetic elements. Belknap Press, Cambridge
Chikhi R, Medvedev P (2013) Informed and automated k-mer size selection for genome assembly. Bioinformatics 30:31–37
Ciomborowska J, Rosikiewicz W, Szklarczyk D, Makałowski W, Makałowska I (2013) “Orphan” retrogenes in the human genome. Mol Biol Evol 30:384–396
Clark FE, Conte MA, Ferreira-Bravo IA, Poletto AP, Martins C, Kocher TD (2017) Dynamic sequence evolution of a sex-associated B chromosome in Lake Malawi cichlid fish. J Hered 108(1):53–62
Drummond AJ, Ashton B, Cheung M et al (2009) Geneious v4.8.5. Available from http://www.geneious.com
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 9:113
Everaert C, Luypaert M, Maag J et al (2017) Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci Rep 7:1559
Fantinatti BEA, Martins C (2016) Development of chromosomal markers based on next-generation sequencing: the B chromosome of the cichlid fish Astatotilapia latifasciata as a model. BMC Genet 17:119
Fantinatti BEA, Mazzuchelli J, Valente GT, Cabral-de-Mello DC, Martins C (2011) Genomic content and new insights on the origin of the B chromosome of the cichlid fish Astatotilapia latifasciata. Genetica 139:1273–1282
Finn RD, Mistry J, Tate J et al (2010) The Pfam protein families database. Nucleic Acids Res 38:D211–D222
Flabet M, Bueno M, Potrzebowski L, Kaessmann H (2009) Evolutionary origin and functions of retrogene introns. Mol Biol Evol 26:2147–2156
Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224
Huang C-J, Lin W-Y, Chang C-M, Choo K-B (2009) Transcription of the rat testis-specific Rtdpoz-T1 and -T2 retrogenes during embryo development: co-transcription and frequent exonisation of transposable element sequences. BMC Mol Biol 10:74
Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP et al (2012) Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep 1:167–178
Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J (2005) Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110:462–467
Kabza M, Ciomborowska J, Makałowska I (2014) RetrogeneDB—a database of animal retrogenes. Mol Biol Evol 31:1646–1648
Kim DY, Kwak E, Kim SH, Lee KH, Woo KC et al (2011) hnRNP Q mediates a phase-dependent translation-coupled mRNA decay of mouse Period3. Nucleic Acids Res 39:8901–8914
Kocher TD (2004) Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet 5:288–298
Kuroiwa A, Terai Y, Kobayashi N et al (2014) Construction of chromosome markers from the Lake Victoria cichlid Paralabidochromis chilotes and their application to comparative mapping. Cytogenet Genome Res 142:112–120
Langmead B, Salzberg S (2012) Fast gapped-read alignment with Bowtie 2. Nature Meth 9:357–359
Li H, Handsaker B, Wysoker A et al (2009) 1000 genome project data processing subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN (2010) RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26:493–500
Lim I, Jung Y, Kim DY, Kim KT (2016) HnRNP Q has a suppressive role in the translation of mouse cryptochrome 1. PLoS One 8:e0159018
Martins C, Cabral-de-Mello DC, Valente GT, Mazzuchelli J, Oliveira SG, Pinhal D (2011) Animal genomes under the focus of cytogenetics. Nova Science Publisher, Hauppauge
Mazzuchelli J, Yang F, Kocher TD, Martins C (2011) Comparative cytogenetic mapping of Sox2 and Sox14 in cichlid fishes and inferences on the genomic organization of both genes in vertebrates. Chromosom Res 19:657–667
McKenna A, Hanna M, Banks E et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), New Orleans, pp 1–8
Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D (2010) Tablet—next generation sequence assembly visualization. Bioinformatics 26:401–402
Muller PY, Janovjak H, Miserez AR, Dobbie Z (2002) Processing of gene expression data generated by quantitative real-time RT-PCR. BioTechniques 32:1372–1374
O’Neill RS, Clark DV (2013) Evolution of three parent genes and their retrogene copies in Drosophila species. Int J Evol Biol 2013:693085
Pan D, Zhang L (2009) Burst of young retrogenes and independent retrogene formation in mammals. PLoS One 4:e5040
Pang AL, Peacock S, Johnson W, Bear DH, Rennert OM, Chan W (2009) Cloning, characterization, and expression analysis of the novel acetyltransferase retrogene Ard1b in the mouse. Biol Reprod 81:302–309
Poletto AB, Ferreira IA, Cabral-de-Mello DC, Nakajima RT, Mazzuchelli J, Ribeiro HB, Venere PC, NirchioM KTD, Martins C (2010a) Chromosome differentiation patterns during cichlid fish evolution. BMC Genet 11:50
Poletto AB, Ferreira IA, Martins C (2010b) The B chromosome of the cichlid fish Haplochromis obliquidens harbors 18S rRNA genes. BMC Genet 11:1
Quaresma AJ, Bressan GC, Gava LM, Lanza DC, Ramos CH, Kobarg J (2009) Human hnRNP Q re-localizes to cytoplasmic granules upon PMA, thapsigargin, arsenite and heat-shock treatments. Exp Cell Res 315:968–980
Quinlan AR (2014) BEDTools: the swiss-army tool for genome feature analysis. In Current protocols in bioinformatics. Curr Protoc Bioinformatics 47:11.12.1–11.12.34
Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66
Ramos E, Cardoso AL, Brown J, Marques DF, Fantinatti BEA, Cabral-de-Mello DC, Martins C (2017) The repetitive DNA element BncDNA, enriched in the B chromosome of the cichlid fish Astatotilapia latifasciata, transcribes a potentially noncoding RNA. Chromosoma 126:313–323
Sambrook J, Russel DW (2001) Molecular cloning. A laboratory manual, 3rd ed. Cold Spring
Shao R, Wang X, Weijdegård B, Norström A, Fernandez-Rodriguez J, Brännström M, Billig H (2012) Coordinate regulation of heterogeneous nuclear ribonucleoprotein dynamics by steroid hormones in the human fallopian tube and endometrium in vivo and in vitro. Am J Physiol Endocrinol Metab 302:1269–1282
Simon P (2003) Q-gene: processing quantitative real-time RT-PCR data. Bioinformatics 19:1439–1440
Valente GT, Conte MA, Fantinatti BEA et al (2014) Origin and evolution of B chromosomes in the cichlid fish Astatotilapia latifasciata based on integrated genomic analyses. Mol Biol Evol 31:2061–2072
Valente GT, Nakajima RT, Fantinatti BEA, Marques DF, Almeida RO, Simões RF, Martins C (2017) B chromosomes: from cytogenetics to systems biology. Chromosoma 126:73–81
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P et al (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Wu TD, Watanabe CK (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2:1859–1875
Xia X (2013) DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol 30:1720–1728
Yoshida K, Terai Y, Mizoiri S et al (2011) B chromosomes have a functional effect on female sex determination in Lake Victoria cichlid fishes. PLoS Genet 7:e1002203
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829
Acknowledgments
This work was financially supported through grants from the São Paulo Research Foundation (FAPESP) (2011/03807-7; 2013/04533-3; 2015/16661-1) and the National Counsel of Technological and Scientific Development (CNPq) (474684/2013-0; 305321/2015-3).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
The experimental procedure was conducted according to the international guidelines of Sao Paulo State University and approved through the Institutional Animal Care and Use Committee (IACUC) (protocol no. 486-2013-CEEA/IBB/UNESP). The animals were euthanized via immersion in a water bath containing benzocaine at 250 mg/l for 10 min.
Consent for publication
Not applicable.
Availability of data and material
All raw data are available in SaciBase (http://sacibase.ibb.unesp.br/).
Competing interests
The authors declare that they have no competing interests.
Additional information
Responsible Editor: Fengtang Yang.
Rights and permissions
About this article
Cite this article
Carmello, B.O., Coan, R.L.B., Cardoso, A.L. et al. The hnRNP Q-like gene is retroinserted into the B chromosomes of the cichlid fish Astatotilapia latifasciata . Chromosome Res 25, 277–290 (2017). https://doi.org/10.1007/s10577-017-9561-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-017-9561-0