Abstract
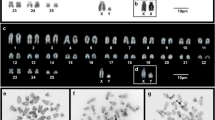
The number of rodent species examined by modern comparative genomic approaches, particularly chromosome painting, is limited. The use of human whole-chromosome painting probes to detect regions of homology in the karyotypes of the rodent index species, the mouse and rat, has been hindered by the highly rearranged nature of their genomes. In contrast, recent studies have demonstrated that non-murid rodents display more conserved genomes, underscoring their suitability for comparative genomic and higher-order systematic studies. Here we provide the first comparative chromosome maps between human and representative rodents of three major rodent lineages Castoridae, Pedetidae and Dipodidae. A comprehensive analysis of these data and those published for Sciuridae show (1) that Castoridae, Pedetidae and Dipodidae form a monophyletic group, and (2) that the European beaver Castor fiber (Castoridae) and the birch mouse Sicista betulina (Dipodidae) are sister species to the exclusion of the springhare Pedetes capensis (Pedetidae), thus resolving an enduring trifurcation in rodent higher-level systematics. Our results together with published data on the Sciuridae allow the formulation of a putative rodent ancestral karyotype (2n = 50) that is thought to comprise the following 26 human chromosomal segments and/or segmental associations: HSA1pq, 1q/10p, 2pq, 2q, 3a, 3b/19p, 3c/21, 4b, 5, 6, 7a, 7b/16p, 8p/4a/8p, 8q, 9/11, 10q, 12a/22a, 12b/22b, 13, 14/15, 16q/19q, 17, 18, 20, X and Y. These findings provide insights into the likely composition of the ancestral rodent karyotype and an improved understanding of placental genome evolution.
Similar content being viewed by others
References
Adkins RM, Walton AH, Honeycutt RL (2003) Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Mol Phylogenet Evol 26: 409–420.
Aniskin VM, Benazzou T, Biltueva L, Dobigny G, Granjon L, Volobouev V (2006) Unusually extensive karyotype reorganization in four congeneric Gerbillus species (Muridae: Gerbillinae). Cytogenet Genome Res 112: 131–140.
Biltueva LS, Graphodatsky AS, Robinson TJ (2006) Pedetes capensis. In: O’Brien SJ, Nash WG, Menninger JC, eds. Atlas of Mammalian Karyotypes. New Jersey: Wiley, p. 310.
Britton-Davidian J, Catalan J, da Graca Ramalhinho M et al. (2000) Rapid chromosomal evolution in island mice. Nature 403: 158.
Bourque G, Tesler G, Pevzner PA (2006) The convergence of cytogenetics and rearrangement-based models for ancestral genome reconstruction. Genome Res 16: 311–313.
Carleton MD, Musser GG (2006) Order Rodentia. In: Wilson DE, Reeder DA, ed. Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd edn. Baltimore, MD: The Johns Hopkins University Press, pp. 745–1600.
Chantry-Darmon C, Bertaud M, Urien C et al. (2005) Expanded comparative mapping between man and rabbit and detection of a new conserved segment between HSA22 and OCU4. Cytogenet Genome Res 111: 134–139.
Couturier J, Dutrillaux B (1986). Chromosomal evolution in Carnivora. Mammalia 50: 124–163.
DeBry RW (2003) Identifying conflicting signal in a multigene analysis reveals a highly resolved tree: the phylogeny of Rodentia (Mammalia). Syst Biol 52: 604–617.
Dobigny G, Ducroz JF, Robinson TJ, Volobouev V (2004) Cytogenetics and cladistics. Syst Biol 53: 470–484.
Dobigny G, Aniskin V, Granjon L, Cornette R, Volobouev V (2005) Recent radiation in West African Taterillus (Rodentia, Gerbillinae): the concerted role of chromosome and climatic changes. Heredity 95: 358–368.
Dutrillaux B, Couturier J (1983) The ancestral karyotype of Carnivora: comparison with that of platyrrhine monkeys. Cytogenet Cell Genet 35: 200–208.
Dutrillaux B, Viegas-Pequignot E, Couturier J (1980) Tres grande analogie de marquage chromosomique entre le lapin (Oryctolagus cunuculus) et les promates dont l’homme. Ann Genet 23: 22–25.
Froenicke L (2005) Origins of primate chromosomes—as delineated by Zoo-FISH and alignments of human and mouse draft genome sequences. Cytogenet Genome Res 108: 122–138.
Froenicke L, Caldes MG, Graphodatsky A et al. (2006) Are molecular cytogenetics and bioinformatics suggesting contradictory models of ancestral mammalian genomes? Genome Res 16: 306–310.
Graphodatsky AS (2006) Castor fiber. In: O’Brien SJ, Nash WG, Menninger JC, eds. Atlas of Mammalian Karyotypes. New Jersey: Wiley, p. 182.
Hayes H, Rogel-Gaillard C, Zijlstra C et al. (2002) Establishment of an R-banded rabbit karyotype nomenclature by FISH localization of 23 chromosme-specific genes on both G- and R-banded chromosomes. Cytogenet Genome Res 98: 199–205.
Huchon D, Madsen O, Sibbald MJJB et al. (2002) Rodent phylogeny and a timescale for the evolution of Glires: evidence from an extensive taxon sampling using three nuclear genes. Mol Biol Evol 19: 1053–1065.
King M (1993) Chromosomal Speciation Revisited (Again). Species Evolution. The Role of Chromosome Change. Cambridge, UK: Cambridge University Press.
Korstanje R, O’Brien PC, Yang F et al. (1999) Complete homology maps of the rabbit (Oryctolagus cuniculus) and human by reciprocal chromosome painting. Cytogenet Cell Genet 86: 317–322.
Li T, O’Brien PC, Biltueva L, Fu B et al. (2004) Evolution of genome organizations of squirrels (Sciuridae) revealed by cross-species chromosome painting. Chromosome Res 12: 317–333.
Li T, Wang J, Su W, Nie W, Yang F (2006) Karyotypic evolution of the family Sciuridae: inferences from the genome organizations of ground squirrels. Cytogenet Genome Res 112: 270–276.
Mouse Genome Sequencing Consortium (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562.
Murphy WJ, Eizirik E, Johnson WE, Ya PZ, Ryder OA, O’Brien SJ (2001a) Molecular phylogenetics and the origins of placental mammals. Nature 409: 614–618.
Murphy WJ, Stanyon R, O’Brien SJ (2001b) Evolution of mammalian genome organization inferred from comparative gene mapping. Genome Biol 2: 0005.1-0005.8.
Murphy WJ, Pevzner PA, O’Brien SJ (2004) Mammalian phylogenomics comes of age. Trends Genet 20: 631–639.
Murphy WJ, Larkin DM, Everts-van der Wind A et al. (2005) Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309: 613–617.
O’Brien SJ, Menniger JC, Nash WG (2006) Atlas of Mammalian Chromosomes. New Jersey: Wiley.
Petit D, Couturier J, Viegas-Pequignot E, Lombard M, Dutrillaux B (1984) Great degree of homoeology between the ancestral karyotype of squirrels (Rodentia) and that of Primates and Carnivora. Ann Genet 27: 201–212.
Rat Genome Sequencing Project Consortium (2004) Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493–521.
Rens W, Fu B, O’Brien PCM, Ferguson-Smith MA (2006) Cross-species chromosom painting. Nat Protoc 1: 783–790.
Richard F, Messaoudi C, Bonnet-Gamier A, Lombard M, Dutrillaux B (2003a) Highly conserved chromosomes in an Asian squirrel (Menetes berdmorei, Rodentia: Sciuridae) as demonstrated by ZOO-FISH with human probes. Chromosome Res 11: 597–603.
Richard F, Lombard M, Dutrillaux B (2003b) Reconstruction of the ancestral karyotype of eutherian mammals. Chromosome Res 11: 605–618.
Robinson TJ, Fu B, Ferguson-Smith MA, Yang F (2004) Cross-species chromosome painting in the golden mole and elephant shrew: support for the mammalian clades Afrotheria and Afroinsectiphillia but not Afroinsectivora. Proc R Soc Lond B Biol Sci 271: 1477–1484.
Robinson TJ, Ruiz-Herrera A, Froenicke L (2006) Dissecting the mammalian genome—new insights into chromosomal evolution. Trends Genet 22: 297–301.
Rokas A, Holland PWH (2000) Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol 15: 454–459.
Scherthan H, Cremer T, Arnason U, Weier HU, Lima-de-Faria A, Frönicke L (1994) Comparative chromosome painting discloses homologous segments in distantly related mammals. Nat Genet 6: 342–347.
Stanyon R, Stone G, Garcia M, Froenicke L (2003) Reciprocal chromosome painting shows that squirrels, unlike murid rodents, have a highly conserved genome organization. Genomics 82: 245–249.
Swofford DL (1998) Phylogenetic Analysis Using Parsimony, version 4.0. Sunderland, MA: Sinauer Associates.
Telenius H, Pelmear AH, Tunnacliffe A et al. (1992) Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosomes Cancer 4: 257–263.
Volleth M, Heller KG, Pfeiffer RA, Hameister H (2002) A comparative ZOO-FISH analysis in bats elucidates the phylogenetic relationships between Megachiroptera and five microchiropteran families. Chromosome Res 10: 477–497.
Viegas-Pequignot E, Petit D, Benazzou T et al. (1986) Chromosomal evolution in Rodents. Mammalia 50: 164–202.
Ward OG, Graphodatsky AS, Wurster-Hill DH, Eremina (Beklemisheva) VR, Park JP, Yu Q (1991) Cytogenetics of beavers: a case of monobrachial fusions. Genome 34: 324–328.
Wang W, Lan H (2000) Rapid and parallel chromosomal number reductions in muntjac deer inferred from mitochondrial DNA phylogeny. Mol Biol Evol 17: 1326–1333.
Wienberg J (2005) Fluorescence in situ hybridization to chromosomes as a tool to understand human and primate genome evolution. Cytogenet Genome Res 108: 139–160.
Wilson DE, Reeder DM (2005) Mammal Species of the World: a Taxonomic and Geographic Reference. Baltimore, MD: The John Hopkins University Press.
Yang F, Müller S, Just R, Ferguson-Smith MA, Wienberg J (1997) Comparative chromosome painting in mammals: human and the Indian muntjac (Muntiacus muntjak vaginalis). Genomics 39: 396–401.
Yang F, O’Brien PCM, Milne BS et al. (1999) A complete comparative chromosome map for the dog, red fox and human and its integration with canine genetic maps. Genomics 62: 189–202.
Yang F, Alkalaeva EZ, Perelman PL et al. (2003) Reciprocal chromosome painting among human, aardvark, and elephant (superorder Afrotheria) reveals the likely eutherian ancestral karyotype. Proc Natl Acad Sci USA 100: 1062–1066.
Yang F, Graphodatsky AS, Li T et al. (2006) Comparative genome maps of the pangolin, hedgehog, sloth, anteater and human revealed by cross-species chromosome painting: further insight into the ancestral karyotype and genome evolution of eutherian mammals. Chromosome Res 14: 283–296.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure S1
Examples of selected human (HSA) chromosome painting probes hybridized to beaver and springhare chromosomes. (JPEG 293 kb)
Supplementary Figure S2
Examples of selected human (HSA) chromosome painting probes hybridized to birch mouse chromosomes. (JPEG 185 kb)
Supplementary Figure S3
Examples of chipmunk (TSE) chromosome 18 probe (which corresponds to HSA7/16) hybridized to beaver chromosomes. (JPEG 83.1 kb)
Supplementary Table S1
Matrix of chromosomal characters corresponding to presence (1) or absence (0) of segmental associations of human chromosomal fragments (e.g., ‘1/2’ corresponds to the segmental association of HSA(1) and HSA(2)), and to inversions. Note that none of the characters was polarized a priori (i.e. there was no a priori assumption about fusion vs. fission). See text for details. (DOC 91kb)
Rights and permissions
About this article
Cite this article
Graphodatsky, A.S., Yang, F., Dobigny, G. et al. Tracking genome organization in rodents by Zoo-FISH. Chromosome Res 16, 261–274 (2008). https://doi.org/10.1007/s10577-007-1191-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-007-1191-5