Abstract
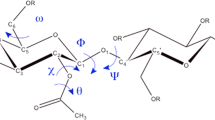
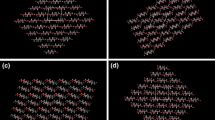
We studied the stability of molecular sheets with four cellotetraoses in an aqueous environment by molecular dynamics simulation to identify the molecular details of first structure as one of the possibilities in the course of crystallization of cellulose I. After simulation, the molecular sheets formed by van der Waals forces along the (11̄0) and (110) crystal plane did not change their structures in an aqueous environment, whereas the other ones formed by hydrogen bonds along the (100) and (200) crystal plane changed into a van der Waals associated molecular sheet, similar to the former. These simulated molecular sheets formed by van der Waals forces were structurally stable in water because of their hydrophilic exterior and hydrophobic interior. Therefore, if the molecular sheet structures are formed in the real system, the sheets formed by van der Waals forces are probably the initial structure of crystallization. A close analysis indicated that these sheets could be classified into two groups in terms of the hydrogen bonding networks, camber angle, and main and side chain conformations. One group was the molecular sheets corresponding to the (110) after simulation. This sheet is probably rigid because intramolecular hydrogen bonds of the chains in the sheet are highly developed. The other group was the molecular sheets corresponding to (200), (100), and (11̄0) crystal plane: the chains in these sheets seemed to be rather flexible due to their moderately developed intramolecular hydrogen bonds.





Similar content being viewed by others
References
Bazooyar F, Momany FA, Bolton K (2012) Validating empirical force fields for molecular-level simulation. Comp Theor Chem 984:119–127
Benziman M, Haigler CH, Brown RM Jr, White AR, Cooper KM (1980) Cellulose biogenesis: polymerization and crystallization are coupled processes in Acetobacter xylinum. Proc Natl Acad Sci USA 77:6678–6682
Bock K, Duus JO (1994) A conformational study of hydroxymethyl groups in carbohydrates investigated by 1H NMR spectroscopy. J Carbohydr Chem 13:513–543
Bosma WB, Appell M, Willett JL, Momany FA (2006) Stepwise hydration of cellobiose by DFT method:1. conformational and structural changes brought about by the addition of one to four water molecules. J. Mol. Struct.:THEOCHEM 776:1–19
Brown RM Jr (1996) The biosynthesis of cellulose. J Macromol Sci Part A Pure Appl Chem 10:1345–1373
Brown RM Jr, Haigler C, Cooper K (1982) Experimental induction of altered nonmicrofibrillar cellulose. Science 218:1141–1142
Cousins SK, Brown RM Jr (1995) Cellulose I microfibril assembly: computational molecular mechanics energy analysis favours bonding by van der Waals forces as the initial step in crystallization. Polymer 36:3885–3888
Delmer DP (1999) Cellulose biosynthesis: exciting times for a difficult field of study 50:245–276
Finkenstadt VL, Millane RP (1998) Crystal structure of valonia cellulose Iβ. Macromolecules 31:7776–7783
French AD (2012) Combining computational chemistry and crystallography for a better understanding of the structure of cellulose. Adv Carbohydr Chem Biochem 67:19–93
French AD, Johnson GP, Cramer CJ, Csonka DI (2012) Conformational analysis of cellobiose by electronic structure theories. Carbohydr Res 350:68–76
Gardner KH, Blackwell J (1974) The structure of native cellulose. Biopolymers 13:1975–2001
Haigler CH, Chanzy H (1988) Electron diffraction analysis of the altered cellulose synthesized by Acetobacter xylinum in the presence of fluorescent brightening agents and direct dyes. J Ultrastruct Mol Struct Res 98:299–311
Hayashi J, Masuda S, Watanabe S (1974) Plane lattice structure in amorphous region of cellulose fibers. Nippon Kagaku Kaishi 5:948–954
Hermans PH (1949) Degree of lateral order in various rayons as deduced from x-ray measurements. J Polym Sci 4:145–151
Horii F, Hirai A, Kitamaru R (1987) In the structures of cellulose: characterization of the solid states. Atalla RH (Ed), American Chemical Society, Washington, DC, pp 119–134
Horii F, Hirai F, Suzuki F (2005) Disordered structure of native cellulose and its production origin. Cellulose Commun 12:179–183
Hu SQ, Gao YG, Tajima K, Sunagawa N, Zhou Y, Kawano S, Fujiwara T, Yoda T, Shimura D, Satoh Y, Munekata M, Tanaka I, Yao M (2010) Structure of bacterial cellulose synthase subunit D octamer with four inner passageways. Proc Natl Acad Sci 107:17957–17961
Kai A, Koseki T (1985) The structure and time evolution of a cellulose sheet in the nascent fibril produced by Acetobacter xylinum. Makromol Chem 186:2609–2614
Kuttel M, Brady JW, Naidoo KJ (2002) Carbohydrate solution simulations: producing a force field with experimentally consistent primary alcohol rotational frequencies and populations. J Comput Chem 23:1236–1243
Langan P, Sukumar N, Nishiyama Y, Chanzy H (2005) Synchrotron x-ray structures of cellulose Iβ and regenerated cellulose II at ambient temperature and 100 K. Cellulose 12:551–562
Miyamoto H, Umemura M, Aoyagi T, Yamane C, Ueda K, Takahashi K (2009) Structural reorganization of molecular sheets derived from cellulose II by molecular dynamics simulations. Carbohydr Res 344:1085–1094
Miyamoto H, Yamane C, Hirase K, Ueda K (2010) Structural rearrangement of chitin molecular sheets in water -Mechanism of structural formation of regenerated chitin proposed by molecular dynamics simulation. J Soc Fiber Sci Technol 66:299–303
Miyamoto H, Ago M, Yamane C, Seguchi M, Ueda K, Okajima K (2011) Supermolecular structure of cellulose/amylose blends prepared from aqueous NaOH solutions and effects of amylose on structural formation of cellulose from its solution. Carbohydr Res 346:807–814
Nishida Y, Ohrui H, Meguro H (1984) 1H-NMR sudies of (6R)- and (6S)-Deuterated D-Hexoses: assignment of the preferred rotamers about C5–C6 bond of d-Glucose and D-Galactose derivatives in solutions. Tetrahedron Lett 25:1575–1578
Nishiyama Y, Langan P, Chanzy H (2002) Crystal structure and hydrogen-bonding system in cellulose Iβ from synchrotron x-ray and neutron fiber diffraction. J Am Chem Soc 124:9074–9082
Sarko A, Muggli R (1974) Packing analysis of carbohydrates and polysaccharides. III. Valonia cellulose and cellulose II. Macromolecules 7:486–494
Saxena IM, Kudlicka K, Okuda K, Brown RM Jr (1994) Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J Bacteriol 176:5735–5752
Schnupf U, Momany FA (2011) Rapidly calculated DFT relaxed iso-potential φ/ψ maps: β-cellobiose. Cellulose 18:887–959
Sun H (1998) COMPASS: an Ab Initio forcefield optimized for condensed-phase application-overview with details on alkane and benzene compounds. J Phys Chem B 102:7338–7364
Acknowledgments
This work was supported by Grant-in-Aid from Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Miyamoto, H., Yamane, C. & Ueda, K. Structural changes in the molecular sheets along (hk0) planes derived from cellulose Iβ by molecular dynamics simulations. Cellulose 20, 1089–1098 (2013). https://doi.org/10.1007/s10570-013-9915-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10570-013-9915-5