Abstract
Purpose
Novel, effective, and safe preventive therapy targets for AF are still needed. Circulating proteins with causal genetic evidence are promising candidates. We aimed to systematically screen circulating proteins for AF drug targets and determine their safety and efficacy using genetic methods.
Methods
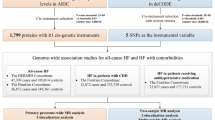
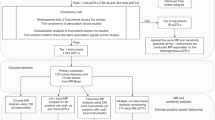
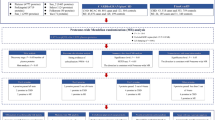
The protein quantitative trait loci (pQTL) of up to 1949 circulating proteins were retrieved from nine large genome-proteome-wide association studies. Two-sample Mendelian Randomization (MR) and colocalization analyses were used to estimate the causal effects of proteins on the risk of AF. Furthermore, phenome-wide MR was conducted to depict side effects and the drug-target databases were searched for drug validation and repurposing.
Results
Systematic MR screen identified 30 proteins as promising AF drug targets. Genetically predicted 12 proteins increased AF risk (TES, CFL2, MTHFD1, RAB1A, DUSP13, SRL, ANXA4, NEO1, FKBP7, SPON1, LPA, MANBA); 18 proteins decreased AF risk (PMVK, UBE2F, SYT11, CHMP3, PFKM, FBP1, TNFSF12, CTSZ, QSOX2, ALAD, EFEMP1, FLRT2, LRIG1, OLA1, SH3BGRL3, IL6R, B3GNT8, FCGR2A). DUSP13 and TNFSF12 possess strong colocalization evidence. For the proteins that were identified, extended phe-MR analysis was conducted to assess their side-effect profiles, while drug-target databases provided information on their approved or investigated indications.
Conclusion
We identified 30 circulating proteins as potential preventive targets for AF.



Similar content being viewed by others
Data Availability
Cis-instruments were queried in the IEU Open GWAS project (https://gwas.mrcieu.ac.uk/). The open-access drug target database (Drugbank: https://go.drugbank.com/; Therapeutic Target Database: http://db.idrblab.net/ttd/; Clinical trial: https://clinicaltrials.gov/) was searched to identify records on past or present clinical drug development programs.
Code availability
Contact the corresponding author for reasonable use.
References
Kornej J, Borschel CS, Benjamin EJ, Schnabel RB. Epidemiology of atrial fibrillation in the 21st century: novel methods and new insights. Circ Res. 2020;127(1):4–20.
Staerk L, Preis SR, Lin H, et al. Protein biomarkers and risk of atrial fibrillation: the FHS. Circ Arrhythm Electrophysiol. 2020;13(2):e007607.
Ko D, Benson MD, Ngo D, et al. Proteomics profiling and risk of new-onset atrial fibrillation: Framingham heart study. J Am Heart Assoc. 2019;8(6):e010976.
Burgess S, Davey Smith G, Davies NM, et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. 2019;4:186.
Lubitz SA, Yin X, Lin HJ, et al. Genetic risk prediction of atrial fibrillation. Circulation. 2017;135(14):1311–20.
Nielsen JB, Thorolfsdottir RB, Fritsche LG, et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet. 2018;50(9):1234–9.
Sun BB, Maranville JC, Peters JE, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558(7708):73–9.
Suhre K, Arnold M, Bhagwat AM, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357.
Folkersen L, Fauman E, Sabater-Lleal M, et al. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet. 2017;13(4):e1006706.
Yao C, Chen G, Song C, et al. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat Commun. 2018;9(1):3268.
Emilsson V, Ilkov M, Lamb JR, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science. 2018;361(6404):769–73.
Zheng J, Haberland V, Baird D, et al. Phenome-wide Mendelian randomization mapping the influence of the plasma proteome on complex diseases. Nat Genet. 2020;52(10):1122–31.
Pietzner M, Wheeler E, Carrasco-Zanini J, et al. Mapping the proteo-genomic convergence of human diseases. Science. 2021;374(6569):eabj1541.
Ferkingstad E, Sulem P, Atlason BA, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet. 2021;53(12):1712–21.
Folkersen L, Gustafsson S, Wang Q, et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab. 2020;2(10):1135–48.
Zhang J, Dutta D, et al. Plasma proteome analyses in individuals of European and African ancestry identify cis-pQTLs and models for proteome-wide association studies. Nat Genet. 2022;54(5):593–602.
Swerdlow DI, Kuchenbaecker KB, Shah S, et al. Selecting instruments for Mendelian randomization in the wake of genome-wide association studies. Int J Epidemiol. 2016;45(5):1600–16.
Hemani G, Zheng J, Elsworth B, et al. The MR-base platform supports systematic causal inference across the human phenome. Elife. 2018;2018:7.
Zuber V, Grinberg NF, Gill D, et al. Combining evidence from Mendelian randomization and colocalization: review and comparison of approaches. Am J Hum Genet. 2022;109(5):767–82.
Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5):e1004383.
Wang Q, Richardson TG, Sanderson E, et al. A phenome-wide bidirectional Mendelian randomization analysis of atrial fibrillation. Int J Epidemiol. 2022;51(4):1153–66.
Ochoa D, Karim M, Ghoussaini M, et al. Human genetics evidence supports two-thirds of the 2021 FDA-approved drugs. Nat Rev Drug Discov. 2022;21(8):551.
Roselli C, Chaffin MD, Weng LC, et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nature genetics. 2018;50(9):1225–33.
Swerdlow DI, Holmes MV, Kuchenbaecker KB, et al. The interleukin-6 receptor as a target for prevention of coronary heart disease: a Mendelian randomisation analysis. Lancet. 2012;379(9822):1214–24.
Meyer MAS, Wiberg S, Grand J, et al. Treatment effects of interleukin-6 receptor antibodies for modulating the systemic inflammatory response after out-of-hospital cardiac arrest (The IMICA Trial): a double-blinded, placebo-controlled, single-center, randomized, clinical trial. Circulation. 2021;143(19):1841–51.
Holmes MV, Ala-Korpela M, Smith GD. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol. 2017;14(10):577–90.
Mutlak M, Kehat I. Dual specific phosphatases (DUSPs) in cardiac hypertrophy and failure. Cell Signal. 2021;84:110033.
Winkles JA. The TWEAK-Fn14 cytokine-receptor axis: discovery, biology and therapeutic targeting. Nat Rev Drug Discov. 2008;7(5):411–25.
Hao L, Ren M, Rong B, et al. TWEAK/Fn14 mediates atrial-derived HL-1 myocytes hypertrophy via JAK2/STAT3 signalling pathway. J Cell Mol Med. 2018;22(9):4344–53.
Zuercher AW, Spirig R, Baz Morelli A, Rowe T, Käsermann F. Next-generation Fc receptor-targeting biologics for autoimmune diseases. Autoimmun Rev. 2019;18(10):102366.
Acknowledgements
This study used summary statistics from large genetic consortia. The authors gratefully acknowledge their contributions to making their datasets publicly available, without which this study would not be possible.
Funding
This work was supported by grants from the Hunan Province Innovative Project (no. 2020SK1013), the National Natural Science Foundation of China (no. 82070356) and the Hunan Provincial Natural Science Foundation of China (no. 2021JJ30033, no. 2021JJ40870).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and software analysis were performed by Zuodong Ning, Yong Zhou and Haochen Lu. The first draft of the manuscript was written by Yunying Huang and all authors commented on previous versions of the manuscript. The validation of data was performed by Feifan Ouyang and the whole project was supervised by Yaozhong Liu. Funding was acquired by Tao Tu and Qiming Liu. All authors read and approved the final manuscript.
Corresponding authors
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
What’s New?
1. Our study systematically screens circulating proteins for AF drug targets and determines their safety and efficacy using genetic methods.
2. We identified 30 proteins as potential preventive drug targets for AF, by using the two-sample MR analysis to evaluate causal roles of 1949 circulating proteins on AF, with a sample size exceeding 1 million. Our finding expands the knowledge regarding the AF prevention targets currently known.
Supplementary information
ESM 1:
Supplementary Table 1. Databases involved in reserch. Supplementary Table 2. The cis-pQTL instuments for 1,949 circulating proteins. Supplementary Table 3. The causal effect of 1,949 circulating proteins on AF risk. Supplementary Table 4. The causal effect of 30 proteins as preventive drug targets on AF. Supplementary Table 5. The colocalization analysis results of the 30 proteins. Supplementary Table 6. The phe-MR analysis results of the 30 proteins. Supplementary Table 7. Potential repurposing opportunities of approved drugs and novel drugs under development.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ning, Z., Huang, Y., Lu, H. et al. Novel Drug Targets for Atrial Fibrillation Identified Through Mendelian Randomization Analysis of the Blood Proteome. Cardiovasc Drugs Ther (2023). https://doi.org/10.1007/s10557-023-07467-8
Accepted:
Published:
DOI: https://doi.org/10.1007/s10557-023-07467-8