Abstract
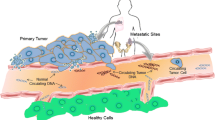
Breast cancer is a spatially and temporally dynamic disease in which differently evolving genetic clones are responsible for progression and clinical outcome. We review tumor heterogeneity and clonal evolution from studies comparing primary tumors and metastasis and discuss plasma circulating tumor DNA as a powerful real-time approach for monitoring the clonal landscape of breast cancer during treatment and recurrence. We found only a few early studies exploring clonal evolution and heterogeneity through analysis of multiregional tissue biopsies of different progression steps in comparison with circulating tumor DNA (ctDNA) from blood plasma. The model of linear progression seemed to be more often reported than the model of parallel progression. The results show complex routes to metastasis, however, and plasma most often reflected metastasis more than primary tumor. The described patterns of evolution and the polyclonal nature of breast cancer have clinical consequences and should be considered during patient diagnosis and treatment selection. Current studies focusing on the relevance of clonal evolution in the clinical setting illustrate the role of liquid biopsy as a noninvasive biomarker for monitoring clonal progression and response to treatment. In the clinical setting, circulating tumor DNA may be an ideal support for tumor biopsies to characterize the genetic landscape of the metastatic disease and to improve longitudinal monitoring of disease dynamics and treatment effectiveness through detection of residual tumor after resection, relapse, or metastasis within a particular patient.


Similar content being viewed by others
References
Ferlay, J. et al. (2018) Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. European Journal of Cancer, 103. Elsevier Ltd, pp. 356–387. https://doi.org/10.1016/j.ejca.2018.07.005.
Harbeck, N., et al. (2019) Breast cancer. Nature Reviews Disease Primers, 5(1). https://doi.org/10.1038/s41572-019-0111-2.
Tellez-Gabriel, M., Knutsen, E., & Perander, M. (2020). Current status of circulating tumor cells, circulating tumor DNA, and exosomes in breast cancer liquid biopsies. International Journal of Molecular Sciences, 21(24):1–23. https://doi.org/10.3390/ijms21249457. (MDPI AG)
Turashvili, G., & Brogi, E. (2014) Tumor heterogeneity in breast cancer. Frontiers in Medicine, 4(DEC):227. https://doi.org/10.3389/FMED.2017.00227.
Joseph, C., Papadaki, A., Althobiti, M., Alsaleem, M., Aleskandarany, M. A., & Rakha, E. A. (2018). Breast cancer intratumour heterogeneity: Current status and clinical implications. Histopathology, 73(5), 717–731. https://doi.org/10.1111/HIS.13642
Rübben, A., & Araujo, A. (2017). Cancer heterogeneity: Converting a limitation into a source of biologic information. Journal of Translational Medicine, 15(1), 1–10. https://doi.org/10.1186/S12967-017-1290-9/FIGURES/3
Klein, C. A. (2009). Parallel progression of primary tumours and metastases. Nature Reviews Cancer, 9(4), 302–312. https://doi.org/10.1038/nrc2627
Torres, L., Ribeiro, F. R., Pandis, N., Andersen, J. A., Heim, S., & Teixeira, M. R. (2007). Intratumor genomic heterogeneity in breast cancer with clonal divergence between primary carcinomas and lymph node metastases. Breast Cancer Research and Treatment, 102(2), 143–155. https://doi.org/10.1007/s10549-006-9317-6
Ellsworth, R. E., Blackburn, H. L., Shriver, C. D., Soon-Shiong, P., & Ellsworth, D. L. (2017). Molecular heterogeneity in breast cancer: State of the science and implications for patient care. Seminars in Cell and Developmental Biology, 64, 65–72. https://doi.org/10.1016/j.semcdb.2016.08.025
Nassar, A., Radhakrishnan, A., Cabrero, I. A., Cotsonis, G. A., & Cohen, C. (2010). Intratumoral heterogeneity of immunohistochemical marker expression in breast carcinoma: A tissue microarray-based study. Applied Immunohistochemistry and Molecular Morphology, 18(5), 433–441. https://doi.org/10.1097/PAI.0b013e3181dddb20
Davis, B. W., Zava, D. T., Locher, G. W., Goldhirsch, A., & Hartmann, W. H. (1984). Receptor heterogeneity of human breast cancer as measured by multiple intratumoral assays of estrogen and progesterone receptor. European Journal of Cancer and Clinical Oncology, 20(3), 375–382. https://doi.org/10.1016/0277-5379(84)90084-1
Appierto, V., di Cosimo, S., Reduzzi, C., Pala, V., Cappelletti, V., & Daidone, M. G. (2017). How to study and overcome tumor heterogeneity with circulating biomarkers: The breast cancer case. Seminars in Cancer Biology, 44(April), 106–116. https://doi.org/10.1016/j.semcancer.2017.04.007
Nik-Zainal, S., et al. (2012). The life history of 21 breast cancers. Cell, 149(5), 994–1007. https://doi.org/10.1016/j.cell.2012.04.023
Baslan, T., et al., (2020) Novel insights into breast cancer copy number genetic heterogeneity revealed by single-cell genome sequencing. eLife, 9:1–21. https://doi.org/10.7554/eLife.51480.
Gerlinger, M., et al. (2012). Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. New England Journal of Medicine, 366(10), 883–892. https://doi.org/10.1056/nejmoa1113205
Martelotto, L. G., Ng, C. K. Y., Piscuoglio, S., Weigelt, B., & Reis-Filho, J. S. (2014). Breast cancer intra-tumor heterogeneity. Breast Cancer Research : BCR, 16(3), 210. https://doi.org/10.1186/BCR3658
Caswell, D. R., & Swanton, C., (2017). The role of tumour heterogeneity and clonal cooperativity in metastasis, immune evasion and clinical outcome. BMC medicine, vol. 15, no. 1. https://doi.org/10.1186/S12916-017-0900-Y.
Marusyk, A., Almendro, V., & Polyak, K. (2012). Intra-tumour heterogeneity: A looking glass for cancer? Nature Reviews Cancer, 12(5):323–334. https://doi.org/10.1038/nrc3261. (Nature Publishing Grou)
Williams, M. J., Sottoriva, A., & Graham, T. A. (2019) Measuring clonal evolution in cancer with genomics, https://doi-org.proxy1-bib.sdu.dk/https://doi.org/10.1146/annurev-genom-083117-021712, vol. 20, pp. 309–329. https://doi.org/10.1146/ANNUREV-GENOM-083117-021712.
Hunter, K. W., Amin, R., Deasy, S., Ha, N. H., & Wakefield, L. (2018). Genetic insights into the morass of metastatic heterogeneity. Nature reviews. Cancer, 18(4), 211. https://doi.org/10.1038/NRC.2017.126
Graham, L. J., et al. (2014). Current approaches and challenges in monitoring treatment responses in breast cancer. Journal of Cancer, 5(1. Jo Cancer):58–68. https://doi.org/10.7150/jca.7047.
Marinovich, M. L., Bernardi, D., Macaskill, P., Ventriglia, A., Sabatino, V., & Houssami, N. (2019). Agreement between digital breast tomosynthesis and pathologic tumour size for staging breast cancer, and comparison with standard mammography. Breast, 43, 59–66. https://doi.org/10.1016/j.breast.2018.11.001
Marinovich, M. L., et al. (2013). Meta-analysis of magnetic resonance imaging in detecting residual breast cancer after neoadjuvant therapy. Journal of the National Cancer Institute, 105(5), 321–333. https://doi.org/10.1093/jnci/djs528
Marinovich, M. L., et al. (2012). Early prediction of pathologic response to neoadjuvant therapy in breast cancer: Systematic review of the accuracy of MRI. Breast, 21(5), 669–677. https://doi.org/10.1016/j.breast.2012.07.006
Pennant, M., et al. (2010). A systematic review of positron emission tomography (PET) and positron emission tomography/computed tomography (PET/CT) for the diagnosis of breast cancer recurrence. Health Technology Assessment, 14(50), 1–120. https://doi.org/10.3310/hta14500
Pinzani, P., et al. (2021). Updates on liquid biopsy: Current trends and future perspectives for clinical application in solid tumors. Clinical chemistry and laboratory medicine, 59(7), 1181–1200. https://doi.org/10.1515/CCLM-2020-1685
Beca, F., & Polyak, K., (2016). Intratumor heterogeneity in breast cancer. Advances in Experimental Medicine and Biology, vol. 882, Springer New York LLC, pp. 169–189. https://doi.org/10.1007/978-3-319-22909-6_7.
Tuaeva, et al. (2019). Translational application of circulating DNA in oncology: Review of the last decades achievements. Cells, 8(10), 1251. https://doi.org/10.3390/cells8101251
Kandoth, C., et al. (2013). Mutational landscape and significance across 12 major cancer types. Nature, 502(7471), 333–339. https://doi.org/10.1038/nature12634
Krøigård, A. B., et al. (2015). Clonal expansion and linear genome evolution through breast cancer progression from pre-invasive stages to asynchronous metastasis. Oncotarget, 6(8):5634–49. https://doi.org/10.18632/oncotarget.3111.
Krøigård, A. B., et al. (2017). Genomic analyses of breast cancer progression reveal distinct routes of metastasis emergence. Scientific Reports, 7, 1–9. https://doi.org/10.1038/srep43813
Krøigård, A. B., et al. (2018). Identification of metastasis driver genes by massive parallel sequencing of successive steps of breast cancer progression. PLoS ONE, 13(1). https://doi.org/10.1371/journal.pone.0189887.
Yates, L. R., et al. (2017). Genomic evolution of breast cancer metastasis and relapse. Cancer Cell, 32(2), 169-184.e7. https://doi.org/10.1016/j.ccell.2017.07.005
Kjällquist, U., et al. (2018). Exome sequencing of primary breast cancers with paired metastatic lesions reveals metastasis-enriched mutations in the A-kinase anchoring protein family (AKAPs). BMC Cancer, 18(1). https://doi.org/10.1186/s12885-018-4021-6.
Ding, L., et al. (2010). Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature, 464(7291), 999–1005. https://doi.org/10.1038/nature08989
Hoadley, K. A., et al. (2016). Tumor evolution in two patients with basal-like breast cancer: A retrospective genomics study of multiple metastases. PLoS Medicine, 13(12). https://doi.org/10.1371/journal.pmed.1002174.
Tang, M.-H. E., et al. (2015). Remarkable similarities of chromosomal rearrangements between primary human breast cancers and matched distant metastases as revealed by whole-genome sequencing. Oncotarget, 6(35):37169–84. https://doi.org/10.18632/oncotarget.5951.
Siegel, M. B., et al. (Apr. 2018). Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. Journal of Clinical Investigation, 128(4), 1371–1383. https://doi.org/10.1172/JCI96153
Paul, M. R., et al. (2020). Genomic landscape of metastatic breast cancer identifies preferentially dysregulated pathways and targets. Journal of Clinical Investigation, 140(8), 4252–4265. https://doi.org/10.1172/JCI129941
Shah, S. P., et al. (2009). Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature, 461(7265), 809–813. https://doi.org/10.1038/nature08489
Ng, C. K. Y., et al. (2017). Genetic heterogeneity in therapy-naïve synchronous primary breast cancers and their metastases. Clinical Cancer Research, 23(15), 4402–4415. https://doi.org/10.1158/1078-0432.CCR-16-3115
P. Savas et al., The subclonal architecture of metastatic breast cancer: Results from a prospective community-based rapid autopsy program ‘CASCADE’. PLoS Medicine, 13(12). https://doi.org/10.1371/journal.pmed.1002204.
Brown, D., et al. (2017). Phylogenetic analysis of metastatic progression in breast cancer using somatic mutations and copy number aberrations. Nature Communications, 8. https://doi.org/10.1038/ncomms14944.
Ullah, I., et al. (2018). Evolutionary history of metastatic breast cancer reveals minimal seeding from axillary lymph nodes. Journal of Clinical Investigation, 128(4), 1355–1370. https://doi.org/10.1172/JCI96149
Blighe, K., et al. (2014). Whole genome sequence analysis suggests intratumoral heterogeneity in dissemination of breast cancer to lymph nodes. PLoS ONE, 9:12. https://doi.org/10.1371/journal.pone.0115346.
My, K., et al. (2009). Tumor self-seeding by circulating cancer cells. Cell, 139(7), 1315–1326. https://doi.org/10.1016/J.CELL.2009.11.025
de Mattos-Arruda, L., et al. (2014). Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: A proof-of-principle. Annals of Oncology, 25(9), 1729–1735. https://doi.org/10.1093/annonc/mdu239
Murtaza, M., et al. (2013). Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature, 497(7447), 108–112. https://doi.org/10.1038/nature12065
Murtaza, M., et al. (2015). Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nature Communications, 6, 4–9. https://doi.org/10.1038/ncomms9760
Butler, T. M., et al. (Aug. 2015). Exome sequencing of cell-free DNA from metastatic cancer patients identifies clinically actionable mutations distinct from primary disease. PLoS ONE, 10(8), e0136407. https://doi.org/10.1371/journal.pone.0136407
Rothé, F., et al. (2014). Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Annals of Oncology, 25(10), 1959–1965. https://doi.org/10.1093/annonc/mdu288
Heidary, M., et al. (2014). The dynamic range of circulating tumor DNA in metastatic breast cancer. Breast Cancer Research, 16(4), 421. https://doi.org/10.1186/s13058-014-0421-y
Koeppel, F., et al. (2017). Whole exome sequencing for determination of tumor mutation load in liquid biopsy from advanced cancer patients. PLoS ONE, 12(11), 1–14. https://doi.org/10.1371/journal.pone.0188174
Yates, L. R., et al. (2015). Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nature Medicine, 21(7), 751–759. https://doi.org/10.1038/nm.3886
Dawson, S. J., et al. (2013). Analysis of circulating tumor DNA to monitor metastatic breast cancer. New England Journal of Medicine, 368(13), 1199–1209. https://doi.org/10.1056/NEJMoa1213261
Frenel, J. S., et al. (2015). Serial next-generation sequencing of circulating cell-free DNA evaluating tumor clone response to molecularly targeted drug administration. Clinical Cancer Research, 21(20), 4586–4596. https://doi.org/10.1158/1078-0432.CCR-15-0584
O’leary, B., et al. (2018). The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discovery, 8(11):1390–1403. https://doi.org/10.1158/2159-8290.CD-18-0264.
Kim, C., et al. (2018). Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell, 173(4), 879-893.e13. https://doi.org/10.1016/j.cell.2018.03.041
Acknowledgements
The authors thank medical writer Claire Gudex for her contribution to editing this review.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kavan, S., Kruse, T.A., Vogsen, M. et al. Heterogeneity and tumor evolution reflected in liquid biopsy in metastatic breast cancer patients: a review. Cancer Metastasis Rev 41, 433–446 (2022). https://doi.org/10.1007/s10555-022-10023-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-022-10023-9