Abstract
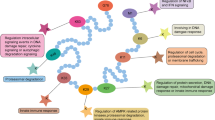
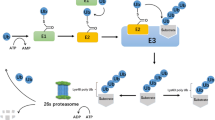
E3 ligases are a class of enzymes that can transfer ubiquitin to substrates for their degradation, which are of importance in cellular homeostasis. Since many oncogenic or tumor-suppressive proteins are reported to be regulated by the ubiquitin-proteasome system (UPS), E3 ligases, which function as substrate interacting modules, have been attracting more and more attention as promising anticancer drug targets due to their pivotal role in conferring substrate specificity. Generally, based on their molecular structure and functional mechanism, E3 ligases can be divided into three major types: homologous to E6-associated protein C-terminus (HECT), really interesting new gene (RING), and RING-in-between-RING (RBR) E3 ligases. Based on the significance of their functions, more bioactive compounds targeting E3 ligases should be developed in the future. In this review, we discuss the important roles of E3 ligases involved in cancer as well as available bioactive compounds targeting various E3 ligases for potential anticancer activity.

Similar content being viewed by others
References
Wilkinson, K. D. (1987). Protein ubiquitination: a regulatory post-translational modification. Anti-Cancer Drug Design, 2(2), 211–229.
Hershko, A. (1983). Ubiquitin: roles in protein modification and breakdown. Cell, 34(1), 11–12.
Liu, J., Shaik, S., Dai, X., Wu, Q., Zhou, X., Wang, Z., et al. (2015). Targeting the ubiquitin pathway for cancer treatment. Biochimica et Biophysica Acta, 1855(1), 50–60. https://doi.org/10.1016/j.bbcan.2014.11.005.
Lipford, J. R., & Deshaies, R. J. (2003). Diverse roles for ubiquitin-dependent proteolysis in transcriptional activation. Nature Cell Biology, 5(10), 845–850. https://doi.org/10.1038/ncb1003-845.
Lianhua, D., Maoliang, R., Zhi, L., Fuzhi, P., & Bin, C. (2016). The role of ubiquitin-proteasome pathway in spermatogenesis. Yi Chuan Hereditas, 38(9), 791–800. https://doi.org/10.16288/j.yczz.16-120.
Chitra, S., Nalini, G., & Rajasekhar, G. (2012). The ubiquitin proteasome system and efficacy of proteasome inhibitors in diseases. International Journal of Rheumatic Diseases, 15(3), 249–260. https://doi.org/10.1111/j.1756-185X.2012.01737.x.
Brown, R., & Kaganovich, D. (2016). Look out autophagy, ubiquilin UPS its game. Cell, 166(4), 797–799. https://doi.org/10.1016/j.cell.2016.07.048.
O’Connell, B. C., & Harper, J. W. (2007). Ubiquitin proteasome system (UPS): what can chromatin do for you? Current Opinion in Cell Biology, 19(2), 206–214. https://doi.org/10.1016/j.ceb.2007.02.014.
Mosesson, Y., Mills, G. B., & Yarden, Y. (2008). Derailed endocytosis: an emerging feature of cancer. Nature Reviews Cancer, 8(11), 835–850. https://doi.org/10.1038/nrc2521.
Okamoto, Y., Ozaki, T., Miyazaki, K., Aoyama, M., Miyazaki, M., & Nakagawara, A. (2003). UbcH10 is the cancer-related E2 ubiquitin-conjugating enzyme. Cancer Research, 63(14), 4167–4173.
Jiang, X., & Chen, Z. J. (2011). The role of ubiquitylation in immune defence and pathogen evasion. Nature Reviews Immunology, 12(1), 35–48. https://doi.org/10.1038/nri3111.
Schulman, B. A., & Harper, J. W. (2009). Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nature Reviews Molecular Cell Biology, 10(5), 319–331. https://doi.org/10.1038/nrm2673.
Bedford, L., Lowe, J., Dick, L. R., Mayer, R. J., & Brownell, J. E. (2011). Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nature Reviews Drug Discovery, 10(1), 29–46. https://doi.org/10.1038/nrd3321.
Ikeda, F., & Dikic, I. (2008). Atypical ubiquitin chains: new molecular signals. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Reports, 9(6), 536–542. https://doi.org/10.1038/embor.2008.93.
Bennett, E. J., & Harper, J. W. (2008). DNA damage: ubiquitin marks the spot. Nature Structural & Molecular Biology, 15(1), 20–22. https://doi.org/10.1038/nsmb0108-20.
Pickart, C. M., & Eddins, M. J. (2004). Ubiquitin: structures, functions, mechanisms. Biochimica et Biophysica Acta, 1695(1–3), 55–72. https://doi.org/10.1016/j.bbamcr.2004.09.019.
Chen, Z. J., & Sun, L. J. (2009). Nonproteolytic functions of ubiquitin in cell signaling. Molecular Cell, 33(3), 275–286. https://doi.org/10.1016/j.molcel.2009.01.014.
Lu, Z., & Hunter, T. (2010). Ubiquitylation and proteasomal degradation of the p21(Cip1), p27(Kip1) and p57(Kip2) CDK inhibitors. Cell Cycle, 9(12), 2342–2352. https://doi.org/10.4161/cc.9.12.11988.
Love, I. M., Shi, D., & Grossman, S. R. (2013). p53 Ubiquitination and proteasomal degradation. Methods in Molecular Biology, 962, 63–73. https://doi.org/10.1007/978-1-62703-236-0_5.
Orlowski, R. Z., & Baldwin Jr., A. S. (2002). NF-kappaB as a therapeutic target in cancer. Trends in Molecular Medicine, 8(8), 385–389.
Huibregtse, J. M., Scheffner, M., Beaudenon, S., & Howley, P. M. (1995). A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proceedings of the National Academy of Sciences of the United States of America, 92(11), 5249.
Rotin, D., & Kumar, S. (2009). Physiological functions of the HECT family of ubiquitin ligases. Nature Reviews Molecular Cell Biology, 10(6), 398–409. https://doi.org/10.1038/nrm2690.
Scheffner, M., & Kumar, S. (2014). Mammalian HECT ubiquitin-protein ligases: biological and pathophysiological aspects. Biochimica et Biophysica Acta, 1843(1), 61–74. https://doi.org/10.1016/j.bbamcr.2013.03.024.
Yang, B., & Kumar, S. (2010). Nedd4 and Nedd4-2: closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death and Differentiation, 17(1), 68–77. https://doi.org/10.1038/cdd.2009.84.
Garcia-Gonzalo, F. R., & Rosa, J. L. (2005). The HERC proteins: functional and evolutionary insights. Cellular and Molecular Life Sciences: CMLS, 62(16), 1826–1838. https://doi.org/10.1007/s00018-005-5119-y.
Nomine, Y., Masson, M., Charbonnier, S., Zanier, K., Ristriani, T., Deryckere, F., et al. (2006). Structural and functional analysis of E6 oncoprotein: insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Molecular Cell, 21(5), 665–678. https://doi.org/10.1016/j.molcel.2006.01.024.
Vander Kooi, C. W., Ohi, M. D., Rosenberg, J. A., Oldham, M. L., Newcomer, M. E., Gould, K. L., et al. (2006). The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases. Biochemistry, 45(1), 121–130. https://doi.org/10.1021/bi051787e.
Morreale, F. E., & Walden, H. (2016). Types of ubiquitin ligases. Cell, 165(1), 248–248 e241. https://doi.org/10.1016/j.cell.2016.03.003.
Leslie, P. L., Ke, H., & Zhang, Y. (2015). The MDM2 RING domain and central acidic domain play distinct roles in MDM2 protein homodimerization and MDM2-MDMX protein heterodimerization. The Journal of Biological Chemistry, 290(20), 12941–12950. https://doi.org/10.1074/jbc.M115.644435.
Cardozo, T., & Pagano, M. (2004). The SCF ubiquitin ligase: insights into a molecular machine. Nature Reviews Molecular Cell Biology, 5(9), 739–751. https://doi.org/10.1038/nrm1471.
Genschik, P., Sumara, I., & Lechner, E. (2013). The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. The EMBO Journal, 32(17), 2307–2320. https://doi.org/10.1038/emboj.2013.173.
Zhao, Y., & Sun, Y. (2013). Cullin-RING ligases as attractive anti-cancer targets. Current Pharmaceutical Design, 19(18), 3215–3225.
Kucharski, T. J., Minshall, P. E., Moustafa-Kamal, M., Turnell, A. S., & Teodoro, J. G. (2017). Reciprocal regulation between 53BP1 and the anaphase-promoting complex/cyclosome is required for genomic stability during mitotic stress. Cell Reports, 18(8), 1982–1995. https://doi.org/10.1016/j.celrep.2017.01.080.
Wenzel, D. M., Lissounov, A., Brzovic, P. S., & Klevit, R. E. (2011). UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature, 474(7349), 105–108. https://doi.org/10.1038/nature09966.
Smit, J. J., & Sixma, T. K. (2014). RBR E3-ligases at work. EMBO Reports, 15(2), 142–154. https://doi.org/10.1002/embr.201338166.
Wang, X., Trotman, L. C., Koppie, T., Alimonti, A., Chen, Z., Gao, Z., et al. (2007). NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell, 128(1), 129–139. https://doi.org/10.1016/j.cell.2006.11.039.
March, H. N., Rust, A. G., Wright, N. A., ten Hoeve, J., de Ridder, J., Eldridge, M., et al. (2011). Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nature Genetics, 43(12), 1202–1209. https://doi.org/10.1038/ng.990.
Martin, E. S., Tonon, G., Sinha, R., Xiao, Y., Feng, B., Kimmelman, A. C., et al. (2007). Common and distinct genomic events in sporadic colorectal cancer and diverse cancer types. Cancer Research, 67(22), 10736–10743. https://doi.org/10.1158/0008-5472.CAN-07-2742.
Eide, P. W., Cekaite, L., Danielsen, S. A., Eilertsen, I. A., Kjenseth, A., Fykerud, T. A., et al. (2013). NEDD4 is overexpressed in colorectal cancer and promotes colonic cell growth independently of the PI3K/PTEN/AKT pathway. Cellular Signalling, 25(1), 12–18. https://doi.org/10.1016/j.cellsig.2012.08.012.
Kim, S. S., Yoo, N. J., Jeong, E. G., Kim, M. S., & Lee, S. H. (2008). Expression of NEDD4-1, a PTEN regulator, in gastric and colorectal carcinomas. APMIS (Acta Pathologica, Microbiologica, et Immunologica Scandinavica), 116(9), 779–784.
Qi, H., Grenier, J., Fournier, A., & Labrie, C. (2003). Androgens differentially regulate the expression of NEDD4L transcripts in LNCaP human prostate cancer cells. Molecular and Cellular Endocrinology, 210(1–2), 51–62.
Booken, N., Gratchev, A., Utikal, J., Weiss, C., Yu, X., Qadoumi, M., et al. (2008). Sezary syndrome is a unique cutaneous T-cell lymphoma as identified by an expanded gene signature including diagnostic marker molecules CDO1 and DNM3. Leukemia, 22(2), 393–399. https://doi.org/10.1038/sj.leu.2405044.
Hu, X. Y., Xu, Y. M., Fu, Q., Yu, J. J., & Huang, J. (2009). Nedd4L expression is downregulated in prostate cancer compared to benign prostatic hyperplasia. European Journal of Surgical Oncology: the Journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology, 35(5), 527–531. https://doi.org/10.1016/j.ejso.2008.09.015.
Zhao, R., Cui, T., Han, C., Zhang, X., He, J., Srivastava, A. K., et al. (2015). DDB2 modulates TGF-beta signal transduction in human ovarian cancer cells by downregulating NEDD4L. Nucleic Acids Research, 43(16), 7838–7849. https://doi.org/10.1093/nar/gkv667.
Qu, M. H., Han, C., Srivastava, A. K., Cui, T., Zou, N., Gao, Z. Q., et al. (2016). miR-93 promotes TGF-beta-induced epithelial-to-mesenchymal transition through downregulation of NEDD4L in lung cancer cells. Tumour Biology, 37(4), 5645–5651. https://doi.org/10.1007/s13277-015-4328-8.
Kito, Y., Bai, J., Goto, N., Okubo, H., Adachi, Y., Nagayama, T., et al. (2014). Pathobiological properties of the ubiquitin ligase Nedd4L in melanoma. International Journal of Experimental Pathology, 95(1), 24–28. https://doi.org/10.1111/iep.12051.
Frampton, A. E., Castellano, L., Colombo, T., Giovannetti, E., Krell, J., Jacob, J., et al. (2014). MicroRNAs cooperatively inhibit a network of tumor suppressor genes to promote pancreatic tumor growth and progression. Gastroenterology, 146(1), 268–277 e218. https://doi.org/10.1053/j.gastro.2013.10.010.
Tanksley, J. P., Chen, X., & Coffey, R. J. (2013). NEDD4L is downregulated in colorectal cancer and inhibits canonical WNT signaling. PLoS One, 8(11), e81514. https://doi.org/10.1371/journal.pone.0081514.
Salah, Z., Melino, G., & Aqeilan, R. I. (2011). Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer Research, 71(5), 2010–2020. https://doi.org/10.1158/0008-5472.CAN-10-3516.
Salah, Z., Itzhaki, E., & Aqeilan, R. I. (2014). The ubiquitin E3 ligase ITCH enhances breast tumor progression by inhibiting the Hippo tumor suppressor pathway. Oncotarget, 5(21), 10886–10900. https://doi.org/10.18632/oncotarget.2540.
Sampath, D., Calin, G. A., Puduvalli, V. K., Gopisetty, G., Taccioli, C., Liu, C. G., et al. (2009). Specific activation of microRNA106b enables the p73 apoptotic response in chronic lymphocytic leukemia by targeting the ubiquitin ligase Itch for degradation. Blood, 113(16), 3744–3753. https://doi.org/10.1182/blood-2008-09-178707.
Massague, J., & Gomis, R. R. (2006). The logic of TGFbeta signaling. FEBS Letters, 580(12), 2811–2820. https://doi.org/10.1016/j.febslet.2006.04.033.
Fukuchi, M., Fukai, Y., Masuda, N., Miyazaki, T., Nakajima, M., Sohda, M., et al. (2002). High-level expression of the Smad ubiquitin ligase Smurf2 correlates with poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Research, 62(24), 7162–7165.
Loukopoulos, P., Shibata, T., Katoh, H., Kokubu, A., Sakamoto, M., Yamazaki, K., et al. (2007). Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinoma: identification of genetic indicators that predict patient outcome. Cancer Science, 98(3), 392–400. https://doi.org/10.1111/j.1349-7006.2007.00395.x.
Fukasawa, H., Yamamoto, T., Fujigaki, Y., Misaki, T., Ohashi, N., Takayama, T., et al. (2010). Reduction of transforming growth factor-beta type II receptor is caused by the enhanced ubiquitin-dependent degradation in human renal cell carcinoma. International Journal of Cancer, 127(7), 1517–1525. https://doi.org/10.1002/ijc.25164.
Jin, C., Yang, Y. A., Anver, M. R., Morris, N., Wang, X., & Zhang, Y. E. (2009). Smad ubiquitination regulatory factor 2 promotes metastasis of breast cancer cells by enhancing migration and invasiveness. Cancer Research, 69(3), 735–740. https://doi.org/10.1158/0008-5472.CAN-08-1463.
Chen, C., Sun, X., Guo, P., Dong, X. Y., Sethi, P., Zhou, W., et al. (2007). Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate cancer. Oncogene, 26(16), 2386–2394. https://doi.org/10.1038/sj.onc.1210021.
Chen, C., Zhou, Z., Ross, J. S., Zhou, W., & Dong, J. T. (2007). The amplified WWP1 gene is a potential molecular target in breast cancer. International Journal of Cancer, 121(1), 80–87. https://doi.org/10.1002/ijc.22653.
Zhang, L., Wu, Z., Ma, Z., Liu, H., Wu, Y., & Zhang, Q. (2015). WWP1 as a potential tumor oncogene regulates PTEN-Akt signaling pathway in human gastric carcinoma. Tumour Biology, 36(2), 787–798. https://doi.org/10.1007/s13277-014-2696-0.
Wu, Z., Zan, P., Li, S., Liu, J., Wang, J., Chen, D., et al. (2015). Knockdown of WWP1 inhibits growth and invasion, but induces apoptosis of osteosarcoma cells. International Journal of Clinical and Experimental Pathology, 8(7), 7869–7877.
Qian, Y. W., Chen, Y., Yang, W., Fu, J., Cao, J., Ren, Y. B., et al. (2012). p28(GANK) prevents degradation of Oct4 and promotes expansion of tumor-initiating cells in hepatocarcinogenesis. Gastroenterology, 142(7), 1547–1558 e1514. https://doi.org/10.1053/j.gastro.2012.02.042.
Yang, R., He, Y., Chen, S., Lu, X., Huang, C., & Zhang, G. (2016). Elevated expression of WWP2 in human lung adenocarcinoma and its effect on migration and invasion. Biochemical and Biophysical Research Communications, 479(2), 146–151. https://doi.org/10.1016/j.bbrc.2016.07.084.
Neumann, M., Vosberg, S., Schlee, C., Heesch, S., Schwartz, S., Gokbuget, N., et al. (2015). Mutational spectrum of adult T-ALL. Oncotarget, 6(5), 2754–2766. https://doi.org/10.18632/oncotarget.2218.
Diouf, B., Cheng, Q., Krynetskaia, N. F., Yang, W., Cheok, M., Pei, D., et al. (2011). Somatic deletions of genes regulating MSH2 protein stability cause DNA mismatch repair deficiency and drug resistance in human leukemia cells. Nature Medicine, 17(10), 1298–1303. https://doi.org/10.1038/nm.2430.
Bonanno, L., Costa, C., Majem, M., Sanchez, J. J., Rodriguez, I., Gimenez-Capitan, A., et al. (2016). Combinatory effect of BRCA1 and HERC2 expression on outcome in advanced non-small-cell lung cancer. BMC Cancer, 16, 312. https://doi.org/10.1186/s12885-016-2339-5.
Narisawa-Saito, M., & Kiyono, T. (2007). Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: roles of E6 and E7 proteins. Cancer Science, 98(10), 1505–1511. https://doi.org/10.1111/j.1349-7006.2007.00546.x.
Gamell, C., Gulati, T., Levav-Cohen, Y., Young, R. J., Do, H., Pilling, P., et al. (2017). Reduced abundance of the E3 ubiquitin ligase E6AP contributes to decreased expression of the INK4/ARF locus in non-small cell lung cancer. Science Signaling, 10(461). https://doi.org/10.1126/scisignal.aaf8223.
Paul, P. J., Raghu, D., Chan, A. L., Gulati, T., Lambeth, L., Takano, E., et al. (2016). Restoration of tumor suppression in prostate cancer by targeting the E3 ligase E6AP. Oncogene, 35(48), 6235–6245. https://doi.org/10.1038/onc.2016.159.
Laurie, N. A., Donovan, S. L., Shih, C. S., Zhang, J., Mills, N., Fuller, C., et al. (2006). Inactivation of the p53 pathway in retinoblastoma. Nature, 444(7115), 61–66. https://doi.org/10.1038/nature05194.
Mayo, L. D., Dixon, J. E., Durden, D. L., Tonks, N. K., & Donner, D. B. (2002). PTEN protects p53 from Mdm2 and sensitizes cancer cells to chemotherapy. The Journal of Biological Chemistry, 277(7), 5484–5489. https://doi.org/10.1074/jbc.M108302200.
Wade, M., Li, Y. C., & Wahl, G. M. (2013). MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nature Reviews Cancer, 13(2), 83–96. https://doi.org/10.1038/nrc3430.
Wang, X. M., Yang, L. Y., Guo, L., Fan, C., & Wu, F. (2009). p53-Induced RING-H2 protein, a novel marker for poor survival in hepatocellular carcinoma after hepatic resection. Cancer, 115(19), 4554–4563. https://doi.org/10.1002/cncr.24494.
Duan, W., Gao, L., Druhan, L. J., Zhu, W. G., Morrison, C., Otterson, G. A., et al. (2004). Expression of Pirh2, a newly identified ubiquitin protein ligase, in lung cancer. Journal of the National Cancer Institute, 96(22), 1718–1721. https://doi.org/10.1093/jnci/djh292.
Logan, I. R., Gaughan, L., McCracken, S. R., Sapountzi, V., Leung, H. Y., & Robson, C. N. (2006). Human PIRH2 enhances androgen receptor signaling through inhibition of histone deacetylase 1 and is overexpressed in prostate cancer. Molecular and Cellular Biology, 26(17), 6502–6510. https://doi.org/10.1128/MCB.00147-06.
Shimada, M., Kitagawa, K., Dobashi, Y., Isobe, T., Hattori, T., Uchida, C., et al. (2009). High expression of Pirh2, an E3 ligase for p27, is associated with low expression of p27 and poor prognosis in head and neck cancers. Cancer Science, 100(5), 866–872.
Marine, J. C. (2012). Spotlight on the role of COP1 in tumorigenesis. Nature Reviews Cancer, 12(7), 455–464. https://doi.org/10.1038/nrc3271.
Dornan, D., Bheddah, S., Newton, K., Ince, W., Frantz, G. D., Dowd, P., et al. (2004). COP1, the negative regulator of p53, is overexpressed in breast and ovarian adenocarcinomas. Cancer Research, 64(20), 7226–7230. https://doi.org/10.1158/0008-5472.CAN-04-2601.
Korphaisarn, K., Morris, V. K., Overman, M. J., Fogelman, D. R., Kee, B. K., Raghav, K. P. S., et al. (2017). FBXW7 missense mutation: a novel negative prognostic factor in metastatic colorectal adenocarcinoma. Oncotarget, 8(24), 39268–39279. https://doi.org/10.18632/oncotarget.16848.
Aydin, I. T., Melamed, R. D., Adams, S. J., Castillo-Martin, M., Demir, A., Bryk, D., et al. (2014). FBXW7 mutations in melanoma and a new therapeutic paradigm. Journal of the National Cancer Institute, 106(6), dju107. https://doi.org/10.1093/jnci/dju107.
Xu, J., Wu, W., Wang, J., Huang, C., Wen, W., Zhao, F., et al. (2017). miR-367 promotes the proliferation and invasion of non-small cell lung cancer via targeting FBXW7. Oncology Reports, 37(2), 1052–1058. https://doi.org/10.3892/or.2016.5314.
Gong, J., Cui, Z., Li, L., Ma, Q., Wang, Q., Gao, Y., et al. (2015). MicroRNA-25 promotes gastric cancer proliferation, invasion, and migration by directly targeting F-box and WD-40 Domain Protein 7, FBXW7. Tumour Biology, 36(10), 7831–7840. https://doi.org/10.1007/s13277-015-3510-3.
Lee, S. W., Li, C. F., Jin, G., Cai, Z., Han, F., Chan, C. H., et al. (2015). Skp2-dependent ubiquitination and activation of LKB1 is essential for cancer cell survival under energy stress. Molecular Cell, 57(6), 1022–1033. https://doi.org/10.1016/j.molcel.2015.01.015.
Hung, W. C., Tseng, W. L., Shiea, J., & Chang, H. C. (2010). Skp2 overexpression increases the expression of MMP-2 and MMP-9 and invasion of lung cancer cells. Cancer Letters, 288(2), 156–161. https://doi.org/10.1016/j.canlet.2009.06.032.
Tosco, P., La Terra Maggiore, G. M., Forni, P., Berrone, S., Chiusa, L., & Garzino-Demo, P. (2011). Correlation between Skp2 expression and nodal metastasis in stage I and II oral squamous cell carcinomas. Oral Diseases, 17(1), 102–108. https://doi.org/10.1111/j.1601-0825.2010.01713.x.
Wood, L. D., Parsons, D. W., Jones, S., Lin, J., Sjoblom, T., Leary, R. J., et al. (2007). The genomic landscapes of human breast and colorectal cancers. Science, 318(5853), 1108–1113. https://doi.org/10.1126/science.1145720.
Gerstein, A. V., Almeida, T. A., Zhao, G., Chess, E., Shih Ie, M., Buhler, K., et al. (2002). APC/CTNNB1 (beta-catenin) pathway alterations in human prostate cancers. Genes, Chromosomes & Cancer, 34(1), 9–16.
Ougolkov, A., Zhang, B., Yamashita, K., Bilim, V., Mai, M., Fuchs, S. Y., et al. (2004). Associations among beta-TrCP, an E3 ubiquitin ligase receptor, beta-catenin, and NF-kappaB in colorectal cancer. Journal of the National Cancer Institute, 96(15), 1161–1170. https://doi.org/10.1093/jnci/djh219.
Muerkoster, S., Arlt, A., Sipos, B., Witt, M., Grossmann, M., Kloppel, G., et al. (2005). Increased expression of the E3-ubiquitin ligase receptor subunit betaTRCP1 relates to constitutive nuclear factor-kappaB activation and chemoresistance in pancreatic carcinoma cells. Cancer Research, 65(4), 1316–1324. https://doi.org/10.1158/0008-5472.CAN-04-1626.
Liu, H., Cheng, E. H., & Hsieh, J. J. (2007). Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes & Development, 21(19), 2385–2398. https://doi.org/10.1101/gad.1574507.
Kato, T., Daigo, Y., Aragaki, M., Ishikawa, K., Sato, M., & Kaji, M. (2012). Overexpression of CDC20 predicts poor prognosis in primary non-small cell lung cancer patients. Journal of Surgical Oncology, 106(4), 423–430. https://doi.org/10.1002/jso.23109.
Wu, W. J., Hu, K. S., Wang, D. S., Zeng, Z. L., Zhang, D. S., Chen, D. L., et al. (2013). CDC20 overexpression predicts a poor prognosis for patients with colorectal cancer. Journal of Translational Medicine, 11, 142. https://doi.org/10.1186/1479-5876-11-142.
Manchado, E., Guillamot, M., de Carcer, G., Eguren, M., Trickey, M., Garcia-Higuera, I., et al. (2010). Targeting mitotic exit leads to tumor regression in vivo: modulation by Cdk1, Mastl, and the PP2A/B55alpha,delta phosphatase. Cancer Cell, 18(6), 641–654. https://doi.org/10.1016/j.ccr.2010.10.028.
Fujita, T., Liu, W., Doihara, H., & Wan, Y. (2008). Regulation of Skp2-p27 axis by the Cdh1/anaphase-promoting complex pathway in colorectal tumorigenesis. The American Journal of Pathology, 173(1), 217–228. https://doi.org/10.2353/ajpath.2008.070957.
Fujita, T., Liu, W., Doihara, H., Date, H., & Wan, Y. (2008). Dissection of the APCCdh1-Skp2 cascade in breast cancer. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 14(7), 1966–1975. https://doi.org/10.1158/1078-0432.CCR-07-1585.
Engelbert, D., Schnerch, D., Baumgarten, A., & Wasch, R. (2008). The ubiquitin ligase APC(Cdh1) is required to maintain genome integrity in primary human cells. Oncogene, 27(7), 907–917. https://doi.org/10.1038/sj.onc.1210703.
Burger, A., Amemiya, Y., Kitching, R., & Seth, A. K. (2006). Novel RING E3 ubiquitin ligases in breast cancer. Neoplasia, 8(8), 689–695. https://doi.org/10.1593/neo.06469.
Burger, A. M., Gao, Y., Amemiya, Y., Kahn, H. J., Kitching, R., Yang, Y., et al. (2005). A novel RING-type ubiquitin ligase breast cancer-associated gene 2 correlates with outcome in invasive breast cancer. Cancer Research, 65(22), 10401–10412. https://doi.org/10.1158/0008-5472.CAN-05-2103.
Ehsani, L., Seth, R., Bacopulos, S., Seth, A., & Osunkoya, A. O. (2013). BCA2 is differentially expressed in renal oncocytoma: an analysis of 158 renal neoplasms. Tumour Biology, 34(2), 787–791. https://doi.org/10.1007/s13277-012-0608-8.
Veeriah, S., Taylor, B. S., Meng, S., Fang, F., Yilmaz, E., Vivanco, I., et al. (2010). Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nature Genetics, 42(1), 77–82. https://doi.org/10.1038/ng.491.
Poulogiannis, G., McIntyre, R. E., Dimitriadi, M., Apps, J. R., Wilson, C. H., Ichimura, K., et al. (2010). PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proceedings of the National Academy of Sciences of the United States of America, 107(34), 15145–15150. https://doi.org/10.1073/pnas.1009941107.
Cesari, R., Martin, E. S., Calin, G. A., Pentimalli, F., Bichi, R., McAdams, H., et al. (2003). Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proceedings of the National Academy of Sciences of the United States of America, 100(10), 5956–5961. https://doi.org/10.1073/pnas.0931262100.
Quinsay, M. N., Lee, Y., Rikka, S., Sayen, M. R., Molkentin, J. D., Gottlieb, R. A., et al. (2010). Bnip3 mediates permeabilization of mitochondria and release of cytochrome c via a novel mechanism. Journal of Molecular and Cellular Cardiology, 48(6), 1146–1156. https://doi.org/10.1016/j.yjmcc.2009.12.004.
Pugh, T. J., Morozova, O., Attiyeh, E. F., Asgharzadeh, S., Wei, J. S., Auclair, D., et al. (2013). The genetic landscape of high-risk neuroblastoma. Nature Genetics, 45(3), 279–284. https://doi.org/10.1038/ng.2529.
Kumar, S., Tomooka, Y., & Noda, M. (1992). Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochemical and Biophysical Research Communications, 185(3), 1155–1161.
Harvey, K. F., & Kumar, S. (1999). Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends in Cell Biology, 9(5), 166–169.
Yang, B., Gay, D. L., MacLeod, M. K., Cao, X., Hala, T., Sweezer, E. M., et al. (2008). Nedd4 augments the adaptive immune response by promoting ubiquitin-mediated degradation of Cbl-b in activated T cells. Nature Immunology, 9(12), 1356–1363. https://doi.org/10.1038/ni.1670.
Ing, B., Shteiman-Kotler, A., Castelli, M., Henry, P., Pak, Y., Stewart, B., et al. (2007). Regulation of Commissureless by the ubiquitin ligase DNedd4 is required for neuromuscular synaptogenesis in Drosophila melanogaster. Molecular and Cellular Biology, 27(2), 481–496. https://doi.org/10.1128/MCB.00463-06.
Fouladkou, F., Lu, C., Jiang, C., Zhou, L., She, Y., Walls, J. R., et al. (2010). The ubiquitin ligase Nedd4-1 is required for heart development and is a suppressor of thrombospondin-1. The Journal of Biological Chemistry, 285(9), 6770–6780. https://doi.org/10.1074/jbc.M109.082347.
Drinjakovic, J., Jung, H., Campbell, D. S., Strochlic, L., Dwivedy, A., & Holt, C. E. (2010). E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron, 65(3), 341–357. https://doi.org/10.1016/j.neuron.2010.01.017.
Uchida, C., & Kitagawa, M. (2016). RING-, HECT-, and RBR-type E3 ubiquitin ligases: involvement in human cancer. Current Cancer Drug Targets, 16(2), 157–174.
Cao, X. R., Lill, N. L., Boase, N., Shi, P. P., Croucher, D. R., Shan, H., et al. (2008). Nedd4 controls animal growth by regulating IGF-1 signaling. Science Signaling, 1(38), ra5. https://doi.org/10.1126/scisignal.1160940.
Araki, N., Umemura, M., Miyagi, Y., Yabana, M., Miki, Y., Tamura, K., et al. (2008). Expression, transcription, and possible antagonistic interaction of the human Nedd4L gene variant: implications for essential hypertension. Hypertension, 51(3), 773–777. https://doi.org/10.1161/HYPERTENSIONAHA.107.102061.
Goel, P., Manning, J. A., & Kumar, S. (2015). NEDD4-2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene, 557(1), 1–10. https://doi.org/10.1016/j.gene.2014.11.051.
Ronzaud, C., Loffing-Cueni, D., Hausel, P., Debonneville, A., Malsure, S. R., Fowler-Jaeger, N., et al. (2013). Renal tubular NEDD4-2 deficiency causes NCC-mediated salt-dependent hypertension. The Journal of Clinical Investigation, 123(2), 657–665. https://doi.org/10.1172/JCI61110.
Gao, S., Alarcon, C., Sapkota, G., Rahman, S., Chen, P. Y., Goerner, N., et al. (2009). Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Molecular Cell, 36(3), 457–468. https://doi.org/10.1016/j.molcel.2009.09.043.
Zhang, Y., Ding, Y., Chen, Y. G., & Tao, Q. (2014). NEDD4L regulates convergent extension movements in Xenopus embryos via Disheveled-mediated non-canonical Wnt signaling. Developmental Biology, 392(1), 15–25. https://doi.org/10.1016/j.ydbio.2014.05.003.
Hustad, C. M., Perry, W. L., Siracusa, L. D., Rasberry, C., Cobb, L., Cattanach, B. M., et al. (1995). Molecular genetic characterization of six recessive viable alleles of the mouse agouti locus. Genetics, 140(1), 255–265.
Lohr, N. J., Molleston, J. P., Strauss, K. A., Torres-Martinez, W., Sherman, E. A., Squires, R. H., et al. (2010). Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. American Journal of Human Genetics, 86(3), 447–453. https://doi.org/10.1016/j.ajhg.2010.01.028.
Bernassola, F., Karin, M., Ciechanover, A., & Melino, G. (2008). The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell, 14(1), 10–21. https://doi.org/10.1016/j.ccr.2008.06.001.
Aki, D., Zhang, W., & Liu, Y. C. (2015). The E3 ligase Itch in immune regulation and beyond. Immunological Reviews, 266(1), 6–26. https://doi.org/10.1111/imr.12301.
Gao, M., Labuda, T., Xia, Y., Gallagher, E., Fang, D., Liu, Y. C., et al. (2004). Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science, 306(5694), 271–275. https://doi.org/10.1126/science.1099414.
Abedini, M. R., Muller, E. J., Brun, J., Bergeron, R., Gray, D. A., & Tsang, B. K. (2008). Cisplatin induces p53-dependent FLICE-like inhibitory protein ubiquitination in ovarian cancer cells. Cancer Research, 68(12), 4511–4517. https://doi.org/10.1158/0008-5472.CAN-08-0673.
Attisano, L., & Wrana, J. L. (2002). Signal transduction by the TGF-beta superfamily. Science, 296(5573), 1646–1647. https://doi.org/10.1126/science.1071809.
David, D., Nair, S. A., & Pillai, M. R. (2013). Smurf E3 ubiquitin ligases at the cross roads of oncogenesis and tumor suppression. Biochimica et Biophysica Acta, 1835(1), 119–128. https://doi.org/10.1016/j.bbcan.2012.11.003.
Nie, J., Xie, P., Liu, L., Xing, G., Chang, Z., Yin, Y., et al. (2010). Smad ubiquitylation regulatory factor 1/2 (Smurf1/2) promotes p53 degradation by stabilizing the E3 ligase MDM2. The Journal of Biological Chemistry, 285(30), 22818–22830. https://doi.org/10.1074/jbc.M110.126920.
Moore, F. E., Osmundson, E. C., Koblinski, J., Pugacheva, E., Golemis, E. A., Ray, D., et al. (2010). The WW-HECT protein Smurf2 interacts with the Docking Protein NEDD9/HEF1 for Aurora A activation. Cell Division, 5, 22. https://doi.org/10.1186/1747-1028-5-22.
Fukunaga, E., Inoue, Y., Komiya, S., Horiguchi, K., Goto, K., Saitoh, M., et al. (2008). Smurf2 induces ubiquitin-dependent degradation of Smurf1 to prevent migration of breast cancer cells. The Journal of Biological Chemistry, 283(51), 35660–35667. https://doi.org/10.1074/jbc.M710496200.
Blank, M., Tang, Y., Yamashita, M., Burkett, S. S., Cheng, S. Y., & Zhang, Y. E. (2012). A tumor suppressor function of Smurf2 associated with controlling chromatin landscape and genome stability through RNF20. Nature Medicine, 18(2), 227–234. https://doi.org/10.1038/nm.2596.
Chen, C., Zhou, Z., Liu, R., Li, Y., Azmi, P. B., & Seth, A. K. (2008). The WW domain containing E3 ubiquitin protein ligase 1 upregulates ErbB2 and EGFR through RING finger protein 11. Oncogene, 27(54), 6845–6855. https://doi.org/10.1038/onc.2008.288.
Zhi, X., & Chen, C. (2012). WWP1: a versatile ubiquitin E3 ligase in signaling and diseases. Cellular and Molecular Life Sciences: CMLS, 69(9), 1425–1434. https://doi.org/10.1007/s00018-011-0871-7.
Chen, W., Jiang, X., & Luo, Z. (2014). WWP2: a multifunctional ubiquitin ligase gene. Pathology Oncology Research, 20(4), 799–803. https://doi.org/10.1007/s12253-014-9838-y.
Soond, S. M., & Chantry, A. (2011). Selective targeting of activating and inhibitory Smads by distinct WWP2 ubiquitin ligase isoforms differentially modulates TGFbeta signalling and EMT. Oncogene, 30(21), 2451–2462. https://doi.org/10.1038/onc.2010.617.
Seto, M. H., Liu, H. L., Zajchowski, D. A., & Whitlow, M. (1999). Protein fold analysis of the B30.2-like domain. Proteins, 35(2), 235–249.
Tae, H., Wei, L., Willemse, H., Mirza, S., Gallant, E. M., Board, P. G., et al. (2011). The elusive role of the SPRY2 domain in RyR1. Channels, 5(2), 148–160.
Garcia-Gonzalo, F. R., Cruz, C., Munoz, P., Mazurek, S., Eigenbrodt, E., Ventura, F., et al. (2003). Interaction between HERC1 and M2-type pyruvate kinase. FEBS Letters, 539(1–3), 78–84.
Utine, G. E., Taskiran, E. Z., Kosukcu, C., Karaosmanoglu, B., Guleray, N., Dogan, O. A., et al. (2017). HERC1 mutations in idiopathic intellectual disability. European Journal of Medical Genetics, 60(5), 279–283. https://doi.org/10.1016/j.ejmg.2017.03.007.
Rosa, J. L., Casaroli-Marano, R. P., Buckler, A. J., Vilaro, S., & Barbacid, M. (1996). p619, a giant protein related to the chromosome condensation regulator RCC1, stimulates guanine nucleotide exchange on ARF1 and Rab proteins. The EMBO Journal, 15(16), 4262–4273.
Sanchez-Tena, S., Cubillos-Rojas, M., Schneider, T., & Rosa, J. L. (2016). Functional and pathological relevance of HERC family proteins: a decade later. Cellular and Molecular Life Sciences: CMLS, 73(10), 1955–1968. https://doi.org/10.1007/s00018-016-2139-8.
Kang, T. H., Lindsey-Boltz, L. A., Reardon, J. T., & Sancar, A. (2010). Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proceedings of the National Academy of Sciences of the United States of America, 107(11), 4890–4895. https://doi.org/10.1073/pnas.0915085107.
Wu, W., Sato, K., Koike, A., Nishikawa, H., Koizumi, H., Venkitaraman, A. R., et al. (2010). HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Research, 70(15), 6384–6392. https://doi.org/10.1158/0008-5472.CAN-10-1304.
Zhu, M., Zhao, H., Liao, J., & Xu, X. (2014). HERC2/USP20 coordinates CHK1 activation by modulating CLASPIN stability. Nucleic Acids Research, 42(21), 13074–13081. https://doi.org/10.1093/nar/gku978.
Bekker-Jensen, S., Rendtlew Danielsen, J., Fugger, K., Gromova, I., Nerstedt, A., Lukas, C., et al. (2010). HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nature Cell Biology, 12(1), 80–86; sup pp 81–12. https://doi.org/10.1038/ncb2008.
Al-Hakim, A. K., Bashkurov, M., Gingras, A. C., Durocher, D., & Pelletier, L. (2012). Interaction proteomics identify NEURL4 and the HECT E3 ligase HERC2 as novel modulators of centrosome architecture. Molecular & Cellular Proteomics, 11(6), M111 014233. https://doi.org/10.1074/mcp.M111.014233.
Chan, N. C., den Besten, W., Sweredoski, M. J., Hess, S., Deshaies, R. J., & Chan, D. C. (2014). Degradation of the deubiquitinating enzyme USP33 is mediated by p97 and the ubiquitin ligase HERC2. The Journal of Biological Chemistry, 289(28), 19789–19798. https://doi.org/10.1074/jbc.M114.569392.
Moroishi, T., Yamauchi, T., Nishiyama, M., & Nakayama, K. I. (2014). HERC2 targets the iron regulator FBXL5 for degradation and modulates iron metabolism. The Journal of Biological Chemistry, 289(23), 16430–16441. https://doi.org/10.1074/jbc.M113.541490.
Mancias, J. D., Pontano Vaites, L., Nissim, S., Biancur, D. E., Kim, A. J., Wang, X., et al. (2015). Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. eLife, 4. https://doi.org/10.7554/eLife.10308.
Kang, T. H., Reardon, J. T., & Sancar, A. (2011). Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Research, 39(8), 3176–3187. https://doi.org/10.1093/nar/gkq1318.
Oda, H., Kumar, S., & Howley, P. M. (1999). Regulation of the Src family tyrosine kinase Blk through E6AP-mediated ubiquitination. Proceedings of the National Academy of Sciences of the United States of America, 96(17), 9557–9562.
Kuhne, C., & Banks, L. (1998). E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. The Journal of Biological Chemistry, 273(51), 34302–34309.
Kim, S., Chahrour, M., Ben-Shachar, S., & Lim, J. (2013). Ube3a/E6AP is involved in a subset of MeCP2 functions. Biochemical and Biophysical Research Communications, 437(1), 67–73. https://doi.org/10.1016/j.bbrc.2013.06.036.
Mantovani, F., & Banks, L. (2001). The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene, 20(54), 7874–7887. https://doi.org/10.1038/sj.onc.1204869.
Huibregtse, J. M., Scheffner, M., & Howley, P. M. (1991). A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. The EMBO Journal, 10(13), 4129–4135.
Olivos, D. J., & Mayo, L. D. (2016). Emerging non-canonical functions and regulation by p53: p53 and stemness. International Journal of Molecular Sciences, 17(12). https://doi.org/10.3390/ijms17121982.
Chene, P. (2003). Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nature Reviews Cancer, 3(2), 102–109. https://doi.org/10.1038/nrc991.
Maguire, M., Nield, P. C., Devling, T., Jenkins, R. E., Park, B. K., Polanski, R., et al. (2008). MDM2 regulates dihydrofolate reductase activity through monoubiquitination. Cancer Research, 68(9), 3232–3242. https://doi.org/10.1158/0008-5472.CAN-07-5271.
Brenkman, A. B., de Keizer, P. L., van den Broek, N. J., Jochemsen, A. G., & Burgering, B. M. (2008). Mdm2 induces mono-ubiquitination of FOXO4. PLoS One, 3(7), e2819. https://doi.org/10.1371/journal.pone.0002819.
Balint, E., Bates, S., & Vousden, K. H. (1999). Mdm2 binds p73 alpha without targeting degradation. Oncogene, 18(27), 3923–3929. https://doi.org/10.1038/sj.onc.1202781.
Sehat, B., Andersson, S., Girnita, L., & Larsson, O. (2008). Identification of c-Cbl as a new ligase for insulin-like growth factor-I receptor with distinct roles from Mdm2 in receptor ubiquitination and endocytosis. Cancer Research, 68(14), 5669–5677. https://doi.org/10.1158/0008-5472.CAN-07-6364.
Uchida, C., Miwa, S., Kitagawa, K., Hattori, T., Isobe, T., Otani, S., et al. (2005). Enhanced Mdm2 activity inhibits pRB function via ubiquitin-dependent degradation. The EMBO Journal, 24(1), 160–169. https://doi.org/10.1038/sj.emboj.7600486.
Wallace, M., Worrall, E., Pettersson, S., Hupp, T. R., & Ball, K. L. (2006). Dual-site regulation of MDM2 E3-ubiquitin ligase activity. Molecular Cell, 23(2), 251–263. https://doi.org/10.1016/j.molcel.2006.05.029.
Yu, G. W., Rudiger, S., Veprintsev, D., Freund, S., Fernandez-Fernandez, M. R., & Fersht, A. R. (2006). The central region of HDM2 provides a second binding site for p53. Proceedings of the National Academy of Sciences of the United States of America, 103(5), 1227–1232. https://doi.org/10.1073/pnas.0510343103.
Gajjar, M., Candeias, M. M., Malbert-Colas, L., Mazars, A., Fujita, J., Olivares-Illana, V., et al. (2012). The p53 mRNA-Mdm2 interaction controls Mdm2 nuclear trafficking and is required for p53 activation following DNA damage. Cancer Cell, 21(1), 25–35. https://doi.org/10.1016/j.ccr.2011.11.016.
Candeias, M. M., Malbert-Colas, L., Powell, D. J., Daskalogianni, C., Maslon, M. M., Naski, N., et al. (2008). P53 mRNA controls p53 activity by managing Mdm2 functions. Nature Cell Biology, 10(9), 1098–1105. https://doi.org/10.1038/ncb1770.
Yap, D. B., Hsieh, J. K., Chan, F. S., & Lu, X. (1999). mdm2: a bridge over the two tumour suppressors, p53 and Rb. Oncogene, 18(53), 7681–7689. https://doi.org/10.1038/sj.onc.1202954.
Stad, R., Little, N. A., Xirodimas, D. P., Frenk, R., van der Eb, A. J., Lane, D. P., et al. (2001). Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Reports, 2(11), 1029–1034. https://doi.org/10.1093/embo-reports/kve227.
Marine, J. C. (2011). MDM2 and MDMX in cancer and development. Current Topics in Developmental Biology, 94, 45–75. https://doi.org/10.1016/B978-0-12-380916-2.00003-6.
Pant, V., Xiong, S., Iwakuma, T., Quintas-Cardama, A., & Lozano, G. (2011). Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proceedings of the National Academy of Sciences of the United States of America, 108(29), 11995–12000. https://doi.org/10.1073/pnas.1102241108.
Sheng, Y., Laister, R. C., Lemak, A., Wu, B., Tai, E., Duan, S., et al. (2008). Molecular basis of Pirh2-mediated p53 ubiquitylation. Nature Structural & Molecular Biology, 15(12), 1334–1342. https://doi.org/10.1038/nsmb.1521.
Leng, R. P., Lin, Y., Ma, W., Wu, H., Lemmers, B., Chung, S., et al. (2003). Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell, 112(6), 779–791.
Li, Q., Lin, S., Wang, X., Lian, G., Lu, Z., Guo, H., et al. (2009). Axin determines cell fate by controlling the p53 activation threshold after DNA damage. Nature Cell Biology, 11(9), 1128–1134.
Hakem, A., Bohgaki, M., Lemmers, B., Tai, E., Salmena, L., Matysiak-Zablocki, E., et al. (2011). Role of Pirh2 in mediating the regulation of p53 and c-Myc. PLoS Genetics, 7(11), e1002360. https://doi.org/10.1371/journal.pgen.1002360.
Jung, Y. S., Qian, Y., & Chen, X. (2011). The p73 tumor suppressor is targeted by Pirh2 RING finger E3 ubiquitin ligase for the proteasome-dependent degradation. The Journal of Biological Chemistry, 286(41), 35388–35395. https://doi.org/10.1074/jbc.M111.261537.
Jung, Y. S., Hakem, A., Hakem, R., & Chen, X. (2011). Pirh2 E3 ubiquitin ligase monoubiquitinates DNA polymerase eta to suppress translesion DNA synthesis. Molecular and Cellular Biology, 31(19), 3997–4006. https://doi.org/10.1128/MCB.05808-11.
Wang, H., Kang, D., Deng, X. W., & Wei, N. (1999). Evidence for functional conservation of a mammalian homologue of the light-responsive plant protein COP1. Current Biology: CB, 9(13), 711–714.
Wei, W., & Kaelin Jr., W. G. (2011). Good COP1 or bad COP1? In vivo veritas. The Journal of Clinical Investigation, 121(4), 1263–1265. https://doi.org/10.1172/JCI57080.
Migliorini, D., Bogaerts, S., Defever, D., Vyas, R., Denecker, G., Radaelli, E., et al. (2011). Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. The Journal of Clinical Investigation, 121(4), 1329–1343. https://doi.org/10.1172/JCI45784.
Uddin, S., Bhat, A. A., Krishnankutty, R., Mir, F., Kulinski, M., & Mohammad, R. M. (2016). Involvement of F-BOX proteins in progression and development of human malignancies. Seminars in Cancer Biology, 36, 18–32. https://doi.org/10.1016/j.semcancer.2015.09.008.
Wang, Z., Liu, P., Inuzuka, H., & Wei, W. (2014). Roles of F-box proteins in cancer. Nature Reviews Cancer, 14(4), 233–247. https://doi.org/10.1038/nrc3700.
Zheng, N., Wang, Z., & Wei, W. (2016). Ubiquitination-mediated degradation of cell cycle-related proteins by F-box proteins. The International Journal of Biochemistry & Cell Biology, 73, 99–110. https://doi.org/10.1016/j.biocel.2016.02.005.
Nash, P., Tang, X., Orlicky, S., Chen, Q., Gertler, F. B., Mendenhall, M. D., et al. (2001). Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature, 414(6863), 514–521. https://doi.org/10.1038/35107009.
Welcker, M., Orian, A., Jin, J., Grim, J. E., Harper, J. W., Eisenman, R. N., et al. (2004). The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proceedings of the National Academy of Sciences of the United States of America, 101(24), 9085–9090. https://doi.org/10.1073/pnas.0402770101.
Nateri, A. S., Riera-Sans, L., Da Costa, C., & Behrens, A. (2004). The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science, 303(5662), 1374–1378. https://doi.org/10.1126/science.1092880.
Koepp, D. M., Schaefer, L. K., Ye, X., Keyomarsi, K., Chu, C., Harper, J. W., et al. (2001). Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science, 294(5540), 173–177. https://doi.org/10.1126/science.1065203.
Mao, J. H., Kim, I. J., Wu, D., Climent, J., Kang, H. C., DelRosario, R., et al. (2008). FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science, 321(5895), 1499–1502. https://doi.org/10.1126/science.1162981.
Tetzlaff, M. T., Yu, W., Li, M., Zhang, P., Finegold, M., Mahon, K., et al. (2004). Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw7 F-box protein. Proceedings of the National Academy of Sciences of the United States of America, 101(10), 3338–3345. https://doi.org/10.1073/pnas.0307875101.
Inuzuka, H., Shaik, S., Onoyama, I., Gao, D., Tseng, A., Maser, R. S., et al. (2011). SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature, 471(7336), 104–109. https://doi.org/10.1038/nature09732.
Murray, A. W. (2004). Recycling the cell cycle: cyclins revisited. Cell, 116(2), 221–234.
Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J. W., et al. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell, 86(2), 263–274.
Weissman, A. M. (2001). Themes and variations on ubiquitylation. Nature Reviews Molecular Cell Biology, 2(3), 169–178. https://doi.org/10.1038/35056563.
Tsvetkov, L. M., Yeh, K. H., Lee, S. J., Sun, H., & Zhang, H. (1999). p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Current Biology: CB, 9(12), 661–664.
Bornstein, G., Bloom, J., Sitry-Shevah, D., Nakayama, K., Pagano, M., & Hershko, A. (2003). Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. The Journal of Biological Chemistry, 278(28), 25752–25757. https://doi.org/10.1074/jbc.M301774200.
Kamura, T., Hara, T., Kotoshiba, S., Yada, M., Ishida, N., Imaki, H., et al. (2003). Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proceedings of the National Academy of Sciences of the United States of America, 100(18), 10231–10236. https://doi.org/10.1073/pnas.1831009100.
Nakayama, K., Nagahama, H., Minamishima, Y. A., Matsumoto, M., Nakamichi, I., Kitagawa, K., et al. (2000). Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. The EMBO Journal, 19(9), 2069–2081. https://doi.org/10.1093/emboj/19.9.2069.
Yu, Z. K., Gervais, J. L., & Zhang, H. (1998). Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proceedings of the National Academy of Sciences of the United States of America, 95(19), 11324–11329.
Zhu, L. (2010). Skp2 knockout reduces cell proliferation and mouse body size: and prevents cancer? Cell Research, 20(6), 605–607. https://doi.org/10.1038/cr.2010.71.
Zheng, N., Zhou, Q., Wang, Z., & Wei, W. (2016). Recent advances in SCF ubiquitin ligase complex: clinical implications. Biochimica et Biophysica Acta, 1866(1), 12–22. https://doi.org/10.1016/j.bbcan.2016.05.001.
Yaron, A., Hatzubai, A., Davis, M., Lavon, I., Amit, S., Manning, A. M., et al. (1998). Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature, 396(6711), 590–594. https://doi.org/10.1038/25159.
Nakayama, K., Hatakeyama, S., Maruyama, S., Kikuchi, A., Onoe, K., Good, R. A., et al. (2003). Impaired degradation of inhibitory subunit of NF-kappa B (I kappa B) and beta-catenin as a result of targeted disruption of the beta-TrCP1 gene. Proceedings of the National Academy of Sciences of the United States of America, 100(15), 8752–8757. https://doi.org/10.1073/pnas.1133216100.
Ding, Q., He, X., Hsu, J. M., Xia, W., Chen, C. T., Li, L. Y., et al. (2007). Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Molecular and Cellular Biology, 27(11), 4006–4017. https://doi.org/10.1128/MCB.00620-06.
Dehan, E., Bassermann, F., Guardavaccaro, D., Vasiliver-Shamis, G., Cohen, M., Lowes, K. N., et al. (2009). betaTrCP- and Rsk1/2-mediated degradation of BimEL inhibits apoptosis. Molecular Cell, 33(1), 109–116. https://doi.org/10.1016/j.molcel.2008.12.020.
Dorrello, N. V., Peschiaroli, A., Guardavaccaro, D., Colburn, N. H., Sherman, N. E., & Pagano, M. (2006). S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science, 314(5798), 467–471. https://doi.org/10.1126/science.1130276.
Tan, M., Gallegos, J. R., Gu, Q., Huang, Y., Li, J., Jin, Y., et al. (2006). SAG/ROC-SCF beta-TrCP E3 ubiquitin ligase promotes pro-caspase-3 degradation as a mechanism of apoptosis protection. Neoplasia, 8(12), 1042–1054. https://doi.org/10.1593/neo.06568.
Spencer, E., Jiang, J., & Chen, Z. J. (1999). Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes & Development, 13(3), 284–294.
Soond, S. M., Townsend, P. A., Barry, S. P., Knight, R. A., Latchman, D. S., & Stephanou, A. (2008). ERK and the F-box protein betaTRCP target STAT1 for degradation. The Journal of Biological Chemistry, 283(23), 16077–16083. https://doi.org/10.1074/jbc.M800384200.
Kudo, Y., Guardavaccaro, D., Santamaria, P. G., Koyama-Nasu, R., Latres, E., Bronson, R., et al. (2004). Role of F-box protein betaTrcp1 in mammary gland development and tumorigenesis. Molecular and Cellular Biology, 24(18), 8184–8194. https://doi.org/10.1128/MCB.24.18.8184-8194.2004.
Belaidouni, N., Peuchmaur, M., Perret, C., Florentin, A., Benarous, R., & Besnard-Guerin, C. (2005). Overexpression of human beta TrCP1 deleted of its F box induces tumorigenesis in transgenic mice. Oncogene, 24(13), 2271–2276. https://doi.org/10.1038/sj.onc.1208418.
Chang, L., Zhang, Z., Yang, J., McLaughlin, S. H., & Barford, D. (2015). Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature, 522(7557), 450–454. https://doi.org/10.1038/nature14471.
Zhou, Z., He, M., Shah, A. A., & Wan, Y. (2016). Insights into APC/C: from cellular function to diseases and therapeutics. Cell Division, 11, 9. https://doi.org/10.1186/s13008-016-0021-6.
Boekhout, M., & Wolthuis, R. (2015). Nek2A destruction marks APC/C activation at the prophase-to-prometaphase transition by spindle-checkpoint-restricted Cdc20. Journal of Cell Science, 128(8), 1639–1653. https://doi.org/10.1242/jcs.163279.
Voets, E., & Wolthuis, R. (2015). MASTL promotes cyclin B1 destruction by enforcing Cdc20-independent binding of cyclin B1 to the APC/C. Biology Open, 4(4), 484–495. https://doi.org/10.1242/bio.201410793.
Peters, J. M. (2002). The anaphase-promoting complex: proteolysis in mitosis and beyond. Molecular Cell, 9(5), 931–943.
Visintin, R., Craig, K., Hwang, E. S., Prinz, S., Tyers, M., & Amon, A. (1998). The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Molecular Cell, 2(6), 709–718.
Huang, J. N., Park, I., Ellingson, E., Littlepage, L. E., & Pellman, D. (2001). Activity of the APC(Cdh1) form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. The Journal of Cell Biology, 154(1), 85–94.
Littlepage, L. E., & Ruderman, J. V. (2002). Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes & Development, 16(17), 2274–2285. https://doi.org/10.1101/gad.1007302.
Stewart, S., & Fang, G. (2005). Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1. Cancer Research, 65(19), 8730–8735. https://doi.org/10.1158/0008-5472.CAN-05-1500.
Stewart, S., & Fang, G. (2005). Anaphase-promoting complex/cyclosome controls the stability of TPX2 during mitotic exit. Molecular and Cellular Biology, 25(23), 10516–10527. https://doi.org/10.1128/MCB.25.23.10516-10527.2005.
Lindon, C., & Pines, J. (2004). Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. The Journal of Cell Biology, 164(2), 233–241. https://doi.org/10.1083/jcb.200309035.
Gutierrez, G. J., Tsuji, T., Chen, M., Jiang, W., & Ronai, Z. A. (2010). Interplay between Cdh1 and JNK activity during the cell cycle. Nature Cell Biology, 12(7), 686–695. https://doi.org/10.1038/ncb2071.
Li, L., Zhou, Y., Wang, G. F., Liao, S. C., Ke, Y. B., Wu, W., et al. (2011). Anaphase-promoting complex/cyclosome controls HEC1 stability. Cell Proliferation, 44(1), 1–9. https://doi.org/10.1111/j.1365-2184.2010.00712.x.
Donzelli, M., Squatrito, M., Ganoth, D., Hershko, A., Pagano, M., & Draetta, G. F. (2002). Dual mode of degradation of Cdc25 A phosphatase. The EMBO Journal, 21(18), 4875–4884.
Bashir, T., Dorrello, N. V., Amador, V., Guardavaccaro, D., & Pagano, M. (2004). Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature, 428(6979), 190–193. https://doi.org/10.1038/nature02330.
Wang, Q., Moyret-Lalle, C., Couzon, F., Surbiguet-Clippe, C., Saurin, J. C., Lorca, T., et al. (2003). Alterations of anaphase-promoting complex genes in human colon cancer cells. Oncogene, 22(10), 1486–1490. https://doi.org/10.1038/sj.onc.1206224.
Amemiya, Y., Azmi, P., & Seth, A. (2008). Autoubiquitination of BCA2 RING E3 ligase regulates its own stability and affects cell migration. Molecular Cancer Research, 6(9), 1385–1396. https://doi.org/10.1158/1541-7786.MCR-08-0094.
Connor, M. K., Azmi, P. B., Subramaniam, V., Li, H., & Seth, A. (2005). Molecular characterization of ring finger protein 11. Molecular Cancer Research, 3(8), 453–461. https://doi.org/10.1158/1541-7786.MCR-04-0166.
Burger, A. M., Kona, F., Amemiya, Y., Gao, Y., Bacopulos, S., & Seth, A. K. (2010). Role of the BCA2 ubiquitin E3 ligase in hormone responsive breast cancer. The Open Cancer Journal, 3(1), 116–123.
Mizuno, K., Kitamura, A., & Sasaki, T. (2003). Rabring7, a novel Rab7 target protein with a RING finger motif. Molecular Biology of the Cell, 14(9), 3741–3752. https://doi.org/10.1091/mbc.E02-08-0495.
Miyakawa, K., Ryo, A., Murakami, T., Ohba, K., Yamaoka, S., Fukuda, M., et al. (2009). BCA2/Rabring7 promotes tetherin-dependent HIV-1 restriction. PLoS Pathogens, 5(12), e1000700. https://doi.org/10.1371/journal.ppat.1000700.
Bacopulos, S., Amemiya, Y., Yang, W., Zubovits, J., Burger, A., Yaffe, M., et al. (2012). Effects of partner proteins on BCA2 RING ligase activity. BMC Cancer, 12, 63. https://doi.org/10.1186/1471-2407-12-63.
Buac, D., Kona, F. R., Seth, A. K., & Dou, Q. P. (2013). Regulation of metformin response by breast cancer associated gene 2. Neoplasia, 15(12), 1379–1390.
Pollak, M. N. (2012). Investigating metformin for cancer prevention and treatment: the end of the beginning. Cancer Discovery, 2(9), 778–790. https://doi.org/10.1158/2159-8290.CD-12-0263.
Hu, Q., & Wang, G. (2016). Mitochondrial dysfunction in Parkinson’s disease. Translational Neurodegeneration, 5, 14. https://doi.org/10.1186/s40035-016-0060-6.
Wang, X., Winter, D., Ashrafi, G., Schlehe, J., Wong, Y. L., Selkoe, D., et al. (2011). PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell, 147(4), 893–906. https://doi.org/10.1016/j.cell.2011.10.018.
Gegg, M. E., Cooper, J. M., Chau, K. Y., Rojo, M., Schapira, A. H., & Taanman, J. W. (2010). Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Human Molecular Genetics, 19(24), 4861–4870. https://doi.org/10.1093/hmg/ddq419.
Bertolin, G., Ferrando-Miguel, R., Jacoupy, M., Traver, S., Grenier, K., Greene, A. W., et al. (2013). The TOMM machinery is a molecular switch in PINK1 and PARK2/PARKIN-dependent mitochondrial clearance. Autophagy, 9(11), 1801–1817. https://doi.org/10.4161/auto.25884.
Geisler, S., Holmstrom, K. M., Skujat, D., Fiesel, F. C., Rothfuss, O. C., Kahle, P. J., et al. (2010). PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature Cell Biology, 12(2), 119–131. https://doi.org/10.1038/ncb2012.
Perez, F. A., & Palmiter, R. D. (2005). Parkin-deficient mice are not a robust model of parkinsonism. Proceedings of the National Academy of Sciences of the United States of America, 102(6), 2174–2179. https://doi.org/10.1073/pnas.0409598102.
Fujiwara, M., Marusawa, H., Wang, H. Q., Iwai, A., Ikeuchi, K., Imai, Y., et al. (2008). Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene, 27(46), 6002–6011. https://doi.org/10.1038/onc.2008.199.
Zhang, C., Lin, M., Wu, R., Wang, X., Yang, B., Levine, A. J., et al. (2011). Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proceedings of the National Academy of Sciences of the United States of America, 108(39), 16259–16264. https://doi.org/10.1073/pnas.1113884108.
Gong, Y., Zack, T. I., Morris, L. G., Lin, K., Hukkelhoven, E., Raheja, R., et al. (2014). Pan-cancer genetic analysis identifies PARK2 as a master regulator of G1/S cyclins. Nature Genetics, 46(6), 588–594. https://doi.org/10.1038/ng.2981.
Vassilev, L. T., Vu, B. T., Graves, B., Carvajal, D., Podlaski, F., Filipovic, Z., et al. (2004). In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science, 303(5659), 844–848. https://doi.org/10.1126/science.1092472.
Ray-Coquard, I., Blay, J. Y., Italiano, A., Le Cesne, A., Penel, N., Zhi, J., et al. (2012). Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. The Lancet Oncology, 13(11), 1133–1140. https://doi.org/10.1016/S1470-2045(12)70474-6.
Ding, Q., Zhang, Z., Liu, J. J., Jiang, N., Zhang, J., Ross, T. M., et al. (2013). Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. Journal of Medicinal Chemistry, 56(14), 5979–5983. https://doi.org/10.1021/jm400487c.
Issaeva, N., Bozko, P., Enge, M., Protopopova, M., Verhoef, L. G., Masucci, M., et al. (2004). Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nature Medicine, 10(12), 1321–1328. https://doi.org/10.1038/nm1146.
Bykov, V. J., Issaeva, N., Shilov, A., Hultcrantz, M., Pugacheva, E., Chumakov, P., et al. (2002). Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nature Medicine, 8(3), 282–288. https://doi.org/10.1038/nm0302-282.
Kitagaki, J., Agama, K. K., Pommier, Y., Yang, Y., & Weissman, A. M. (2008). Targeting tumor cells expressing p53 with a water-soluble inhibitor of Hdm2. Molecular Cancer Therapeutics, 7(8), 2445–2454. https://doi.org/10.1158/1535-7163.MCT-08-0063.
Yang, Y., Ludwig, R. L., Jensen, J. P., Pierre, S. A., Medaglia, M. V., Davydov, I. V., et al. (2005). Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell, 7(6), 547–559. https://doi.org/10.1016/j.ccr.2005.04.029.
Herman, A. G., Hayano, M., Poyurovsky, M. V., Shimada, K., Skouta, R., Prives, C., et al. (2011). Discovery of Mdm2-MdmX E3 ligase inhibitors using a cell-based ubiquitination assay. Cancer Discovery, 1(4), 312–325. https://doi.org/10.1158/2159-8290.CD-11-0104.
Yuan, Y., Liao, Y. M., Hsueh, C. T., & Mirshahidi, H. R. (2011). Novel targeted therapeutics: inhibitors of MDM2, ALK and PARP. Journal of Hematology & Oncology, 4, 16. https://doi.org/10.1186/1756-8722-4-16.
Reed, D., Shen, Y., Shelat, A. A., Arnold, L. A., Ferreira, A. M., Zhu, F., et al. (2010). Identification and characterization of the first small molecule inhibitor of MDMX. The Journal of Biological Chemistry, 285(14), 10786–10796. https://doi.org/10.1074/jbc.M109.056747.
Wang, H., Ma, X., Ren, S., Buolamwini, J. K., & Yan, C. (2011). A small-molecule inhibitor of MDMX activates p53 and induces apoptosis. Molecular Cancer Therapeutics, 10(1), 69–79. https://doi.org/10.1158/1535-7163.MCT-10-0581.
Ling, X., Xu, C., Fan, C., Zhong, K., Li, F., & Wang, X. (2014). FL118 induces p53-dependent senescence in colorectal cancer cells by promoting degradation of MdmX. Cancer Research, 74(24), 7487–7497. https://doi.org/10.1158/0008-5472.CAN-14-0683.
Orlicky, S., Tang, X., Neduva, V., Elowe, N., Brown, E. D., Sicheri, F., et al. (2010). An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nature Biotechnology, 28(7), 733–737. https://doi.org/10.1038/nbt.1646.
Huang, H. L., Weng, H. Y., Wang, L. Q., Yu, C. H., Huang, Q. J., Zhao, P. P., et al. (2012). Triggering Fbw7-mediated proteasomal degradation of c-Myc by oridonin induces cell growth inhibition and apoptosis. Molecular Cancer Therapeutics, 11(5), 1155–1165. https://doi.org/10.1158/1535-7163.MCT-12-0066.
Chan, C. H., Morrow, J. K., Li, C. F., Gao, Y., Jin, G., Moten, A., et al. (2013). Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell, 154(3), 556–568. https://doi.org/10.1016/j.cell.2013.06.048.
Chen, Q., Xie, W., Kuhn, D. J., Voorhees, P. M., Lopez-Girona, A., Mendy, D., et al. (2008). Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood, 111(9), 4690–4699. https://doi.org/10.1182/blood-2007-09-112904.
Roy, S., Kaur, M., Agarwal, C., Tecklenburg, M., Sclafani, R. A., & Agarwal, R. (2007). p21 and p27 induction by silibinin is essential for its cell cycle arrest effect in prostate carcinoma cells. Molecular Cancer Therapeutics, 6(10), 2696–2707. https://doi.org/10.1158/1535-7163.MCT-07-0104.
Yang, E. S., & Burnstein, K. L. (2003). Vitamin D inhibits G1 to S progression in LNCaP prostate cancer cells through p27Kip1 stabilization and Cdk2 mislocalization to the cytoplasm. The Journal of Biological Chemistry, 278(47), 46862–46868. https://doi.org/10.1074/jbc.M306340200.
Blees, J. S., Bokesch, H. R., Rubsamen, D., Schulz, K., Milke, L., Bajer, M. M., et al. (2012). Erioflorin stabilizes the tumor suppressor Pdcd4 by inhibiting its interaction with the E3-ligase beta-TrCP1. PLoS One, 7(10), e46567. https://doi.org/10.1371/journal.pone.0046567.
Nakajima, H., Fujiwara, H., Furuichi, Y., Tanaka, K., & Shimbara, N. (2008). A novel small-molecule inhibitor of NF-kappaB signaling. Biochemical and Biophysical Research Communications, 368(4), 1007–1013. https://doi.org/10.1016/j.bbrc.2008.01.166.
Zeng, X., Sigoillot, F., Gaur, S., Choi, S., Pfaff, K. L., Oh, D. C., et al. (2010). Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell, 18(4), 382–395. https://doi.org/10.1016/j.ccr.2010.08.010.
Sackton, K. L., Dimova, N., Zeng, X., Tian, W., Zhang, M., Sackton, T. B., et al. (2014). Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature, 514(7524), 646–649. https://doi.org/10.1038/nature13660.
Jiang, J., Thyagarajan-Sahu, A., Krchnak, V., Jedinak, A., Sandusky, G. E., & Sliva, D. (2012). NAHA, a novel hydroxamic acid-derivative, inhibits growth and angiogenesis of breast cancer in vitro and in vivo. PLoS One, 7(3), e34283. https://doi.org/10.1371/journal.pone.0034283.
Jiang, J., & Sliva, D. (2010). Novel medicinal mushroom blend suppresses growth and invasiveness of human breast cancer cells. International Journal of Oncology, 37(6), 1529–1536.
Jiang, J., Jedinak, A., & Sliva, D. (2011). Ganodermanontriol (GDNT) exerts its effect on growth and invasiveness of breast cancer cells through the down-regulation of CDC20 and uPA. Biochemical and Biophysical Research Communications, 415(2), 325–329. https://doi.org/10.1016/j.bbrc.2011.10.055.
Morrow, J. K., Lin, H. K., Sun, S. C., & Zhang, S. (2015). Targeting ubiquitination for cancer therapies. Future Medicinal Chemistry, 7(17), 2333–2350. https://doi.org/10.4155/fmc.15.148.
Mattern, M. R., Wu, J., & Nicholson, B. (2012). Ubiquitin-based anticancer therapy: carpet bombing with proteasome inhibitors vs surgical strikes with E1, E2, E3, or DUB inhibitors. Biochimica et Biophysica Acta, 1823(11), 2014–2021. https://doi.org/10.1016/j.bbamcr.2012.05.005.
Funding
This work was supported by the National Natural Sciences Foundation of China (81502065 and 81672926), China Postdoctoral Science Foundation Funded Project (2016T90613, 2015M580574, and 2016M592146), Natural Sciences Foundation of Shandong Province (ZR2014HQ009), Shandong Postdoctoral Innovation Project (201602037), Qingdao Innovation Applied Basic Research Project (16-5-1-56-JCH), and Qingdao Postdoctoral Research Project (2015167 and 2015157).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Wang, D., Ma, L., Wang, B. et al. E3 ubiquitin ligases in cancer and implications for therapies. Cancer Metastasis Rev 36, 683–702 (2017). https://doi.org/10.1007/s10555-017-9703-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-017-9703-z