Abstract
Purpose
The recent publication of the ACOSOG Z1031 trial results demonstrated that Ki-67 proliferation marker-based neoadjuvant endocrine therapy response monitoring could be used for tailoring the use of adjuvant chemotherapy in ER+HER2-negative breast cancer patients. In this paper, we describe the development of the Ki-67 clinical trial assay used for this study.
Methods
Ki-67 assay assessment focused on reproducing a 2.7% Ki-67 cut-point (CP) required for calculating the Preoperative Endocrine Prognostic Index and a 10% CP for poor endocrine therapy response identification within the first month of neoadjuvant endocrine treatment. Image analysis was assessed to increase the efficiency of the scoring process. Clinical outcome concordance for two independent Ki-67 scores was the primary performance metric.
Results
Discordant scores led to a triage approach where cases with complex histological features that software algorithms could not resolve were flagged for visual point counting (17%). The final Ki-67 scoring approach was run on T1/2 N0 cases from the P024 and POL trials (N = 58). The percent positive agreement for the 2.7% CP was 87.5% (95% CI 61.7–98.5%); percent negative agreement 88.9% (95% CI: 65.3–98.6%). Minor discordance did not affect the ability to predict similar relapse-free outcomes (Log-Rank P = 0.044 and P = 0.055). The data for the 10% early triage CP in the POL trial were similar (N = 66), the percentage positive agreement was 100%, and percent negative agreement 93.55% (95% CI: 78.58–99.21%). The independent survival predictions were concordant (Log-rank P = 0.0001 and P = 0.01).
Conclusions
We have developed an efficient and reproducible Ki-67 scoring system that was approved by the Clinical Trials Evaluation Program for NCI-supported neoadjuvant endocrine therapy trials. Using the methodology described here, investigators are able to identify a subgroup of patients with ER+HER2-negative breast cancer that can be safely managed without the need of adjuvant chemotherapy.
Similar content being viewed by others
Introduction
Biomarkers of cell proliferation are used to assess prognosis and response to cancer treatment, and most clinical assays are based on Ki-67 immunohistochemistry (IHC) [1]. The Ki-67 nuclear protein is present in proliferating cells but absent in cells in G0 [2]. For breast cancer, Ki-67 analysis is relevant for estrogen receptor positive (ER+) early stage breast cancer [3,4,5] which presents as a spectrum of tumors with clinically indolent (Luminal A) or more aggressive features (Luminal B) [6]. While the “luminal” classification is based on gene expression analysis, a Ki-67 cut-point of 14% of cells staining positive has been proposed as a surrogate for the distinction between luminal A and luminal B [7]. This cut-point was considered clinically useful by the St. Gallen breast cancer consensus panel [8] but the concerns of the American Society of Clinical Oncology Tumor Marker Guideline Committee regarding the lack of rigor in Ki-67 scoring algorithms and the questionable validity of decision-making cut-points has slowed clinical implementation [9].
Ki-67 analysis also has potential for monitoring endocrine therapy response, which requires testing a tumor specimen after endocrine treatment has been initiated, for example, in surgical specimens after neoadjuvant aromatase inhibition [10]. The independent prognostic value of on-treatment Ki-67 was combined with pathologic tumor stage and ER status to develop the preoperative endocrine prognostic index (PEPI). A PEPI score of 0 (pT1/2N0, Ki-67 ≤2.7% and persistently expressed ER) was associated with such favorable long-term outcome after neoadjuvant endocrine therapy in the P024 trial [11] and IMPACT trial [10, 12] that chemotherapy was proposed to be unnecessary [13].
Recently, Ellis et al. published long-term follow-up results of the ACOSOG Z1031 trial in which clinical decisions were based on the PEPI score [14]. In ACOSOG Z1031 Cohort B, the authors tested the hypothesis that Ki-67-based algorithms can also address the concern that patients who are poorly responsive to neoadjuvant endocrine therapy should ideally be identified early for triage to alternate treatment, such as neoadjuvant chemotherapy or immediate surgery. The authors also successfully identified a subgroup of patients, based on PEPI scores that could be safely spared from adjuvant chemotherapy. In this paper, we describe the validation of Ki-67 cut-points relevant to neoadjuvant endocrine treatment monitoring and the development and validation of the Ki-67 clinical trial assay for prospective studies, used in ACOSOG Z1031 trial [14].
Methods
Database analysis for early Ki-67 cut-point for early triage to alternate treatment
Published data on research use only (RUO) quantitative polymerase chain reaction (qPCR)-based assignments of PAM50 luminal subtype (A vs. B) and RUO Ki-67 data from TMA analysis was made available from six hundred sixty-seven tumors with clinical ER-positive status from University of British Columbia. Of these tumors, 358 were classified as Luminal A and 309 as Luminal B [7]. Published Ki-67 data and clinical outcomes from the IMPACT trial [12] and POL Trial [15, 16] were used for the development of cut-points for prospective validation.
Tumor samples for Ki-67 clinical trial assay development
For training the scanner and image analysis-based Ki-67 quantification algorithm, 61 node-positive samples from the P024 trial were examined. For assay validation for the early triage cut-point, core needle biopsies taken after 4 weeks of neoadjuvant endocrine therapy from 66 patients were accessed [15]. For validation of the 2.7% cut-point required for the PEPI score, surgical specimens from 58 patients with pathological stage 1 or 2A tumors were available from a combination of the POL trial [15] and the P024 trial [11].
Ki-67 assay methodology
The research use only (RUO) Ki-67 assay employed to stain the P024 and POL samples for combined survival analysis employed the SP6 monoclonal antibody (Neomarkers) on a Shandon Sequenza® Immunostainer using published methodology [13]. For the CLIA clinical trial assay, 5 micron sections from POL and P024 trials were subjected to H&E and Ki-67 staining in the CLIA-certified Washington University AMP laboratory using the CONFIRM anti-Ki-67 (30-9) rabbit monoclonal primary antibody as a pre-diluted reagent on a Benchmark XT platform according to the manufacturer instructions (Ventana, Tucson, AZ). Tonsil was used as the assay control.
Ki-67 scoring approaches
For visual point counting (VPC), photomicrographs of three randomly selected fields were taken at 40X with a background grid and color printed (more fields to achieve the minimal cell count). Each observer counted both the total number tumor cells and the number of Ki-67-positive cells that intersect with first grid line. This process is repeated on every third gridline. All the cells on the slide were counted if three fields could not be obtained however at least 200 total tumor cells were required. For Ki-67 image analysis of the CLIA clinical trial assay, slides were scanned with the iScan Coreo scanner (Ventana). The computer image was reviewed and “Areas of Interest” (AOI) were selected at 4X magnification using the following guidelines: (1) identify the largest AOI of representative clear invasive tumor; (2) exclude DCIS, vessels, lymphocytes; (3) avoid AOIs in peri-necrotic or necrotic areas; (4) identify at least 3 AOIs and a maximum of 10. The image analysis was performed using the FDA cleared VENTANA Companion Algorithm Ki-67 (30-9) and the VENTANA VIRTUOSO software (Roche).
Assessment of concordance
Two pathologists, blinded to each other’s data and any data from earlier analyses of the samples, independently reviewed the Ki-67 slide scans and identified AOI for either image analysis or VPC methodology. Similarly blinded trained technicians generated the VPC Ki-67 percentage.
Statistical analysis
Analysis of variance using a scatter-plot analysis was calculated using Pearsons’ correlation and Spearman correlation coefficients. Two pathologist concordance for the 2.7 and 10% cut-points were analyzed using four-by-four contingency table analysis, simple Kappa coefficients and percent positive and negative agreements. The prognostic effect of modified PEPI 0 (pT1/2, N0, Ki-67 ≤ 2.7%) vs. non-0 assignments based on the CLIA Ki-67 assay determined using the Kaplan–Meier method. The log-rank test was conducted to examine statistical significance. Similar analyses were performed to correlate survival outcomes of patients with early on-treatment Ki-67 (>10 vs ≤ 10%) in the POL trial. Bland–Altman plots were generated to assess bias between pathologists.
Results
A Ki-67-based definition of poorly endocrine therapy responsive tumors for triage to alternate treatment
To develop a Ki-67-based approach for the early identification of non-responders within a month of starting treatment, we examined the interaction between baseline Ki-67 levels and a qPCR-PAM50-based definition of luminal A versus luminal B breast cancer using published data [17]. Using ROC methodology, a 10% Ki-67 cut-point of Ki-67 best served as a surrogate for the genomic luminal definitions in this data set (Fig. 1). We therefore hypothesized that tumors with an early Ki-67 value above 10% despite endocrine therapy would be enriched for endocrine therapy resistant, luminal B-type tumors with a high relapse rate. This is supported by the early on-treatment data from the POL [15] and IMPACT [12] trials which indicated that Ki-67 levels >10% predicted a higher level of Ki-67 in the surgical sample, a higher PEPI score, a smaller number of patients in the PEPI-0 group, and worse RFS [14].
PEPI score validation and modification
Long-term outcomes from the POL trial provided an opportunity to further validate of the PEPI score. While the number of cases was modest, no relapses were observed in 10 patients with PEPI 0 tumors after a median follow-up of 59 months (Fig. 2a). We also developed a modified PEPI score that did not include ER status at surgery, because of clinical trial proposals that included the use of the estrogen receptor down-regulator fulvestrant, the use of which confounds the interpretation of ER levels after treatment initiation [18]. In the P024, IMPACT and POL trials, patients with modified PEPI score of 0 were all ER+ (Allred score 3–8) because ER Allred score 0–2 post aromatase inhibitor or tamoxifen treatment was associated with either a high Ki-67 or high tumor staging (or both) excluding these cases from PEPI-0 status without the need for information on ER. In the combined P024 trial/POL trial data, no relapses were observed in the 29 patients (19 pT1N0, 10 pT2N0) with modified PEPI-0 status (i.e., without scoring ER) during a median follow-up of 62.5 months (Fig. 2b).
Validation of visual point counting (VPC) for outcome prediction after neoadjuvant endocrine therapy
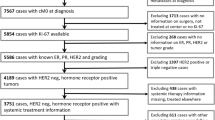
In previous analyses, VPC methodology was routinely used but this approach had not been formally assessed as part of a clinical trial assay. Available surgical tumor samples from pT1/2 N0 cases in the POL and P024 trials were therefore stained using the commercial 30-9 antibody assay in a CLIA-certified laboratory. Stage 1 or 2A cases were chosen because a Ki-67 cut-point (CP) of ≤2.7% is the only factor that determines the modified PEPI score of 0. The REMARK sample flow chart for the duplicate study is provided in Fig. 3a. Outcome predictions were reproducible, with no relapses observed for patients assigned modified PEPI 0 (Ki-67 ≤ 2.7%) status by either pathologist (Fig. 3b). Analysis of Ki-67 as a continuous variable indicated that the Spearman Correlation Coefficient was 0.938 (p < 0.0001) (Figure S2A), and there was no trend for increased discordance across the range of Ki-67 values (Figure S2B). The positive CP agreement was 13/13 (100%). The negative agreement was 9/12 (0.75) (95% exact confidence limit: 0.428–0.945). Simple Kappa Coefficient was 0.7573 (95% Confidence limit: 0.5073; 1) (Table 1, S1A).
Assessment of an image analysis approach for Ki-67 scoring
The performance of VPC, while technically adequate, is laborious and therefore not ideal for real-time clinical reporting. We therefore considered a Ki-67 scoring approach using an FDA cleared scanner and image interpretation software to determine if these tools were appropriate. For training, the 30-9 antibody-based commercial assay was conducted on 61 surgical samples from patients with node-positive disease in the P024 trial (Figure S1). The slides were scanned and then analyzed by two pathologists who independently reviewed the images and drew areas of interest (AOI) for Ki-67 scoring. In five instances, the algorithm did not accurately differentiate between benign and malignant cells. These cases were noteworthy for abundant lymphocyte infiltration, sparse tumor cells where tumor cells were streaming through the tissue with a large amount of intervening stroma, abundant marking of non-fascicular “plump” fibroblasts, or when the Ki-67 stain was generally diffuse and nuclear staining was faint. Excluding these cases, the Spearman Correlation Coefficient was 0.89 (p < 0.0001) (Figure S3A). The Bland–Altman plot showed no bias in scoring between the two pathologists across the range of Ki-67 values (Figure S3B). The CP concordance was then analyzed. For the 2.7% cut-point, the positive agreement was 29/30 (0.96) (95% exact confidence limit: 0.82–0.99). The negative agreement was 23/26 (0.88) (95% exact confidence limit: 0.69–0.97). The kappa coefficient was 0.85 (95% confidence limit: 0.71; 0.99). Using the 10% cut-point, the positive agreement was 100%, and the negative agreement was 46/47 (0.97) (95% exact confidence limit: 0.88–1). The kappa coefficient was 0.93 (95% Confidence limit: 0.81; 1.0) (Table 1, S1B). A “locked-down” scoring standard operating procedure (SOP) was generated that included an option to triage to VPC if the pathology was judged too complex for the scanner to differentiate benign from malignant cells (Fig. 4).
Validation of combined imaging/VPC Ki-67 scoring SOP for the 2.7% Ki-67 cut-point
To validate the combined imaging/VPC SOP for Ki-67 scoring, the CLIA assay stained slides used for the VPC assessment were scanned and independently assessed by two pathologists. The sample flow chart is shown in Fig. 5a. Kaplan–Meier analysis by modified PEPI 0 is shown in Fig. 5b for the two separate scoring exercises. Again, no relapses were observed in patients with modified PEPI 0 during the follow-up using this scoring method from either pathologist. Continuous data analysis indicated that the Spearman Correlation Coefficient was 0.86 (p < 0.0001) (Figure S4A). No scoring bias was observed across the scored range (Figure S4B). The percentage positive agreement between the two pathologists in scoring the 2.7% CP Ki-67 using the SOP was 0.87 (95% CI 0.61–0.98). The negative agreement was 0.88 (95% CI 0.65–0.98). Simple kappa coefficient was 0.76 (95% CI 0.54–0.98) (Table 1, S1C).
Validation of combined imaging/VPC Ki-67 scoring SOP for the 10% Ki-67 cut-point
To validate the combined imaging/VPC approach for the 10% cut-point one-month biopsies from the POL trial were stained using the Ki-67 30-9 clinical trial assay, scanned, and then independently reviewed for algorithm accuracy and independently scored by two pathologists. The REMARK sample flow chart is shown in Fig. 6a. Concordant Kaplan–Meier analyses for the 10% cut-point for two separate scoring exercises are shown in Fig. 6b. The poor outcome for patients in the >10% category was reproducible. The Spearman Correlation Coefficient was 0.86 (p < 0.0001) (Figure S5A). No scoring bias was observed across the scoring range (Figure S5B). The percentage positive agreement between the two pathologists in scoring the 10% CP Ki-67 using the SOP was 100%. The negative agreement was 93.6 (78.6–99.2). The kappa coefficient was 0.86(0.66–1) (Table 1, S1D).
Discussion
We have developed an efficient and reproducible Ki-67 scoring system that was approved by the Clinical Trials Evaluation Program (CTEP) for NCI-supported neoadjuvant endocrine therapy trials. The combination of image analysis with triage to VPC, when deemed necessary, respects the finding that the image analysis software does not always differentiate between certain types of normal and malignant cells. This approach also emphasizes the critical role of the pathologist in the review of the scanned images to determine the most appropriate scoring approach (image analysis or VPC) when the histology is complex. The sample flow charts illustrate that while pathologists may have different interpretations for the requirement for visual point counting, these differences do not strongly affect clinical outcome prediction. The VPC triage rate was, on average 17%, demonstrating that the image analysis approach can be used in the majority of cases, markedly reducing the need to conduct laborious VPC to a manageable number of cases. A weakness of our study is that the sample sets were denuded by earlier analyses and produced very modest sample sizes, and therefore, our analysis did not produce evidence for immediate clinical utility. However, the Ki-67 clinical trial assay developed and described in this paper was further validated in ACOSOG Z1031A study. In that trial, with a median follow-up of 5.5 years, this Ki67 methodology was able to identify a subgroup of patients with PEPI score = 0 (Ki-67 ≤ 2.7%, T1/2, N0) that were safely managed without adjuvant chemotherapy. Among patients with PEPI = 0 score that were managed without chemotherapy, only 4 out of 119 presented with a relapse during follow-up. The triage rate to VPC in the Z1031A trial was 6%, even lower than what we found in the POL and P024 sample sets.
An issue not addressed in the scoring algorithm proposed herein concerns cases where the Ki-67 staining is not uniform—our VPC or image analysis approach requires random fields. We consider a Ki-67 heterogeneity-agnostic approach equivalent to genomic approaches that also do not clearly respect tissue heterogeneity. While analysis of heterogeneity, or “Ki-67 hot spot” analysis, should be pursued, this is a complex problem that will require the development of a “hot-spot” definition that can be shown to drive outcome more effectively than an analysis of all the tumor cells in the section.
Another point of controversy is the Ki-67 cut-point as a surrogate for luminal A versus luminal B breast cancer. In our current analysis, 10% has the best operating characteristics while an earlier publication on a different dataset using similar methodology suggested 14% [7], which suggests a narrow range of values for this purpose. From the perspective of this paper, the 10% cut-point was more conservative and serves the purpose of early identification of patients with luminal B-type tumors with endocrine therapy resistance characteristics well. The rapid onset of advanced disease for patients with Ki-67 > 10% despite aromatase inhibitor therapy (see Fig. 6b for example) underscores the importance of developing a robust clinical trial strategy for this high-risk population.
When we submitted our Ki-67 clinical trial assay to the FDA they ruled the proposed treatment algorithms as “no significant risk” because Ki-67 analysis actually reduces the risk of under-treatment. This conclusion was based on the analysis of chemotherapy use according to PEPI score shows that when medical oncologists rely on pathological stage alone after neoadjuvant endocrine therapy most patients with low stage do not receive chemotherapy. Combined analysis of the P024, IMPACT and POL trials showed that only 8% of patients with pathological stage 1 or 2A disease received adjuvant chemotherapy (Table S2). Thus, the FDA considered that knowledge of the Ki-67 value in the pathological specimen reduced the risk of under-treatment for patients with low pathological stage tumors but aggressive biological characteristics (high on-treatment Ki-67).
Even though ASCO still does not support Ki67 in its clinical guidelines, a recent editorial acknowledges our team’s efforts as “an important step in the direction of clinical respectability for Ki67 as a useful breast cancer prognosticator” [19]. The next necessary step is already being taken as the Ki-67 clinical trial assay we described in this paper is being prospectively validated in the ALTERNATE trial (NCT01953588).
References
Dowsett M, Nielsen TO, A’Hern R, Bartlett J, Coombes RC, Cuzick J et al (2011) Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst 103(22):1656–1664
Gerdes J, Schwab U, Lemke H, Stein H (1983) Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer 31(1):13–20
Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA (2010) Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 11(2):174–183
Urruticoechea A, Smith IE, Dowsett M (2005) Proliferation marker Ki-67 in early breast cancer. J Clin Oncol 23(28):7212–7220
Stuart-Harris R, Caldas C, Pinder SE, Pharoah P (2008) Proliferation markers and survival in early breast cancer: a systematic review and meta-analysis of 85 studies in 32,825 patients. Breast 17(4):323–334
Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T et al (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27(8):1160–1167
Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J et al (2009) Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 101(10):736–750
Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ (2011) Strategies for subtypes–dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol 22(8):1736–1747
Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S et al (2007) American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol 25(33):5287–5312
Dowsett M, Smith IE, Ebbs SR, Dixon JM, Skene A, A’Hern R et al (2007) Prognostic value of Ki67 expression after short-term presurgical endocrine therapy for primary breast cancer. J Natl Cancer Inst 99(2):167–170
Eiermann W, Paepke S, Appfelstaedt J, Llombart-Cussac A, Eremin J, Vinholes J et al (2001) Preoperative treatment of postmenopausal breast cancer patients with letrozole: a randomized double-blind multicenter study. Ann Oncol 12(11):1527–1532
Smith IE, Dowsett M, Ebbs SR, Dixon JM, Skene A, Blohmer JU et al (2005) Neoadjuvant treatment of postmenopausal breast cancer with anastrozole, tamoxifen, or both in combination: the Immediate Preoperative Anastrozole, Tamoxifen, or Combined with Tamoxifen (IMPACT) multicenter double-blind randomized trial. J Clin Oncol 23(22):5108–5116
Ellis MJ, Tao Y, Luo J, A’Hern R, Evans DB, Bhatnagar AS et al (2008) Outcome prediction for estrogen receptor-positive breast cancer based on postneoadjuvant endocrine therapy tumor characteristics. J Natl Cancer Inst 100(19):1380–1388
Ellis MJ, Suman VJ, Hoog J, Goncalves R, Sanati S, Creighton CJ et al (2017) Ki67 proliferation index as a tool for chemotherapy decisions during and after neoadjuvant aromatase inhibitor treatment of breast cancer: results From the American College of Surgeons Oncology Group Z1031 Trial (Alliance). J Clin Oncol. 35(10):1061–1069
Olson JA Jr, Budd GT, Carey LA, Harris LA, Esserman LJ, Fleming GF et al (2009) Improved surgical outcomes for breast cancer patients receiving neoadjuvant aromatase inhibitor therapy: results from a multicenter phase II trial. J Am Coll Surg 208(5):906–914 discussion 15-6
Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW et al (2012) Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486(7403):353–360
Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T et al (2010) A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res 16(21):5222–5232
Robertson JF, Dixon JM, Sibbering DM, Jahan A, Ellis IO, Channon E et al (2013) A randomized trial to assess the biological activity of short-term (pre-surgical) fulvestrant 500 mg plus anastrozole versus fulvestrant 500 mg alone or anastrozole alone on primary breast cancer. Breast Cancer Res 15(2):R18
Sledge GW Jr (2017) Put Some PEPI in Your Step: Ki67’s Long Road to Respectability. J Clin Oncol 35(10):1031–1032
Acknowledgements
We would like to thank Dr Lisa McShane for guidance on the development plan for the Ki-67 assay, Dr Mitch Dowsett and Dr Roger A’Hern for analysis of the IMPACT data and Dr Torsten Nielsen for analysis of the University of British Columbia tamoxifen series. We would also like to thank the FDA for their review on the necessity of an IDE application for Ki-67 analysis in CTEP sponsored neoadjuvant endocrine therapy trials.
Funding
This work was supported by grants to Dr. Matthew Ellis from the AVON Foundation, R01 CA095614, the Barnes-Jewish Foundation, the Breast Cancer Research Foundation and Novartis (for biomarker studies on the P024 trial). The tissue procurement core was supported by NCI P50 CA68438. Dr R. Goncalves was supported by the AVON foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
Dr. Matthew Ellis has stock ownership of Bioclassifier LLC and Prosigna Nanostring. Dr. Matthew Ellis receives consultant fees from AstraZeneca, Novartis and Pfizer. Dr. Michael Barnes is an employee and has stock ownership at Roche/Ventana medical systems. The other authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Goncalves, R., DeSchryver, K., Ma, C. et al. Development of a Ki-67-based clinical trial assay for neoadjuvant endocrine therapy response monitoring in breast cancer. Breast Cancer Res Treat 165, 355–364 (2017). https://doi.org/10.1007/s10549-017-4329-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-017-4329-y