Abstract
Purpose
NSD3 has been implicated as a candidate driver oncogene from the 8p11-p12 locus, and we have previously published evidence for its amplification and overexpression in human breast cancer. This aim of this study was to further characterize the transforming function of NSD3 in vivo.
Methods
We generated a transgenic mouse model in which NSD3 gene expression was driven by the MMTV promoter and expressed in mammary epithelium of FVB mice. Mammary glands were fixed and whole mounts were stained with carmine to visualize gland structure. Mammary tumors were formalin-fixed, and paraffin embedded (FFPE) tumors were stained with hematoxylin and eosin.
Results
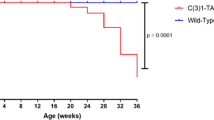
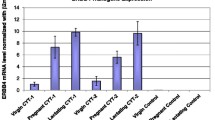
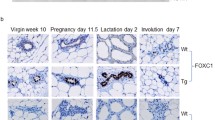
Pups born to transgenic females were significantly underdeveloped compared to pups born to WT females due to a lactation defect in transgenic female mice. Whole mount analysis of the mammary glands of transgenic female mice revealed a profound defect in functional differentiation of mammary gland alveoli that resulted in the lactation defect. We followed parous and virgin NSD3 transgenic and control mice to 50 weeks of age and observed that several NSD3 parous females developed mammary tumors. Whole mount analysis of the mammary glands of tumor-bearing mice revealed numerous areas of mammary hyperplasia and ductal dysplasia. Histological analysis showed that mammary tumors were high-grade ductal carcinomas, and lesions present in other mammary glands exhibited features of alveolar hyperplasia, ductal dysplasia, and carcinoma in situ.
Conclusions
Our results are consistent with our previous studies and demonstrate that NSD3 is a transforming breast cancer oncogene.





Similar content being viewed by others
References
Adelaide J, Chaffanet M, Imbert A, Allione F, Geneix J, Popovici C, van Alewijk D, Trapman J, Zeillinger R, Borresen-Dale AL et al (1998) Chromosome region 8p11-p21: refined mapping and molecular alterations in breast cancer. Genes Chromosomes Cancer 22(3):186–199
Ray ME, Yang ZQ, Albertson D, Kleer CG, Washburn JG, Macoska JA, Ethier SP (2004) Genomic and expression analysis of the 8p11–12 amplicon in human breast cancer cell lines. Cancer Res 64(1):40–47
Bilal E, Vassallo K, Toppmeyer D, Barnard N, Rye IH, Almendro V, Russnes H, Borresen-Dale AL, Levine AJ, Bhanot G et al (2012) Amplified loci on chromosomes 8 and 17 predict early relapse in ER-positive breast cancers. PLoS ONE 7(6):e38575
Gelsi-Boyer V, Orsetti B, Cervera N, Finetti P, Sircoulomb F, Rouge C, Lasorsa L, Letessier A, Ginestier C, Monville F et al (2005) Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res 3(12):655–667
Carapeti M, Aguiar RC, Watmore AE, Goldman JM, Cross NC (1999) Consistent fusion of MOZ and TIF2 in AML with inv (8) (p11q13). Cancer Genet Cytogenet 113(1):70–72
Stec I, den Dunnen JT (2001) WHSC1L1, on human chromosome 8p11.2, closely resembles WHSC1 and maps to a duplicated region shared with 4p16.3. Genomics 76(1–3):5–8
Cully M, Shiu J, Piekorz RP, Muller WJ, Done SJ, Mak TW (2005) Transforming acidic coiled coil 1 promotes transformation and mammary tumorigenesis. Cancer Res 65(22):10363–10370
Elbauomy Elsheikh S, Green AR, Lambros MB, Turner NC, Grainge MJ, Powe D, Ellis IO, Reis-Filho JS (2007) FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res 9(2):R23
Streicher KL, Yang ZQ, Draghici S, Ethier SP (2007) Transforming function of the LSM1 oncogene in human breast cancers with the 8p11-12 amplicon. Oncogene 26(14):2104–2114
Bernard-Pierrot I, Gruel N, Stransky N, Vincent-Salomon A, Reyal F, Raynal V, Vallot C, Pierron G, Radvanyi F, Delattre O (2008) Characterization of the recurrent 8p11-12 amplicon identifies PPAPDC1B, a phosphatase protein, as a new therapeutic target in breast cancer. Cancer Res 68(17):7165–7175
Zhang J, Liu X, Datta A, Govindarajan K, Tam WL, Han J, George J, Wong C, Ramnarayanan K, Phua TY et al (2009) RCP is a human breast cancer-promoting gene with Ras-activating function. J Clin Invest 119(8):2171–2183
Slorach EM, Chou J, Werb Z (2011) Zeppo1 is a novel metastasis promoter that represses E-cadherin expression and regulates p120-catenin isoform expression and localization. Genes Dev 25(5):471–484
Wang G, Liu G, Wang X, Sethi S, Ali-Fehmi R, Abrams J, Zheng Z, Zhang K, Ethier S, Yang ZQ (2012) ERLIN2 promotes breast cancer cell survival by modulating endoplasmic reticulum stress pathways. BMC Cancer 12:225
Turner-Ivey B, Guest ST, Irish JC, Kappler CS, Garrett-Mayer E, Wilson RC, Ethier SP (2014) KAT6A, a chromatin modifier from the 8p11-p12 amplicon is a candidate oncogene in luminal breast cancer. Neoplasia 16(8):644–655
Yang ZQ, Liu G, Bollig-Fischer A, Giroux CN, Ethier SP (2010) Transforming properties of 8p11-12 amplified genes in human breast cancer. Cancer Res 70(21):8487–8497
Irish JC, Mills JN, Turner-Ivey B, Wilson RC, Guest ST, Rutkovsky A, Dombkowski A, Kappler CS, Hardiman G, Ethier SP (2016) Amplification of WHSC1L1 regulates expression and estrogen-independent activation of ERalpha in SUM-44 breast cancer cells and is associated with ERalpha over-expression in breast cancer. Mol Oncol 10:850–865
Angrand PO, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P (2001) NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics 74(1):79–88
Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ et al (2005) High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA 102(27):9625–9630
Taketani T, Taki T, Nakamura H, Taniwaki M, Masuda J, Hayashi Y (2009) NUP98-NSD3 fusion gene in radiation-associated myelodysplastic syndrome with t(8;11) (p11;p15) and expression pattern of NSD family genes. Cancer Genet Cytogenet 190(2):108–112
Zhou Z, Thomsen R, Kahns S, Nielsen AL (2010) The NSD3L histone methyltransferase regulates cell cycle and cell invasion in breast cancer cells. Biochem Biophys Res Commun 398(3):565–570
He C, Li F, Zhang J, Wu J, Shi Y (2013) The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition. J Biol Chem 288(7):4692–4703
Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS, Suzuki Y, Pappin DJ, Joshua-Tor L, Vakoc CR (2015) NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol Cell 60(6):847–859
Chen Y, McGee J, Chen X, Doman TN, Gong X, Zhang Y, Hamm N, Ma X, Higgs RE, Bhagwat SV et al (2014) Identification of druggable cancer driver genes amplified across TCGA datasets. PLoS ONE 9(5):e98293
Luo J, Liu S, Leung S, Gru AA, Tao Y, Hoog J, Ho J, Davies SR, Allred DC, Salavaggione AL et al (2017) An mRNA gene expression-based signature to identify FGFR1-amplified estrogen receptor-positive breast tumors. J Mol Diagn 19(1):147–161
Yang ZQ, Streicher KL, Ray ME, Abrams J, Ethier SP (2006) Multiple interacting oncogenes on the 8p11-p12 amplicon in human breast cancer. Cancer Res 66(24):11632–11643
Cornen S, Guille A, Adelaide J, Addou-Klouche L, Finetti P, Saade MR, Manai M, Carbuccia N, Bekhouche I, Letessier A et al (2014) Candidate luminal B breast cancer genes identified by genome, gene expression and DNA methylation profiling. PLoS ONE 9(1):e81843
Andre F, Bachelot T, Campone M, Dalenc F, Perez-Garcia JM, Hurvitz SA, Turner N, Rugo H, Smith JW, Deudon S et al (2013) Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin Cancer Res 19(13):3693–3702
Pearson A, Smyth E, Babina IS, Herrera-Abreu MT, Tarazona N, Peckitt C, Kilgour E, Smith NR, Geh C, Rooney C et al (2016) High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov 6(8):838–851
Acknowledgements
We would like to specifically thank Michael J. Kern, Ph.D. of the Gene Function Core at the Medical University of South Carolina and all of the technicians for their assistance in generating the NSD3 transgenic mice. We would also like to thank Kiwana Gibbs and Yuan Shao, Ph.D. of the Biorepository and Tissue Analysis Shared Resource at the Medical University of South Carolina for providing histological sections and staining of all mammary glands and tumors.
Authors’ contribution
Conception and design of the study was conducted by SE. Transgenic mice were handled by ES for ear tagging/tail snips, euthanizing, and harvesting of all mammary glands for whole mount preparation. Genotyping and transgenic mouse record keeping for the NSD3 colony was performed by BI. Both ES and BI performed carmine staining of whole mounts, cultured tumor-derived mammary cells, and performed western blotting on lysates obtained from NSD3-derived mammary tumor cells. AR organized mammary glands and generated images for scoring hyperplastic lesions seen in whole mounts. LS is the breast cancer pathologist for the Hollings Cancer Center and along with JM performed histopathologic analysis of mammary lesions and tumors. The manuscript was written by SE, BI, ES, AR, and LS.
Funding
This study was funded by the 2RO1CA100724, NCI 2P30CA138313, and the Chalsty Breast Cancer Research Fund 1K12CA457688-O1A1.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. The Institutional Animal Care and Use Committee (IACUC) at the Medical University of SC (MUSC) approved all animal experiments, AR#3397. Male and female WT FVB/N mice were bred under typical conditions with unlimited access to food and water. Animals were euthanized per IACUC approved protocol during all experiments. This article does not contain any studies with human participants performed by any of the authors.
Additional information
Brittany Turner-Ivey and Ericka L. Smith have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Turner-Ivey, B., Smith, E.L., Rutkovsky, A.C. et al. Development of mammary hyperplasia, dysplasia, and invasive ductal carcinoma in transgenic mice expressing the 8p11 amplicon oncogene NSD3. Breast Cancer Res Treat 164, 349–358 (2017). https://doi.org/10.1007/s10549-017-4258-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-017-4258-9