Abstract
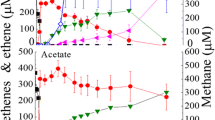
The dechlorinating Dehalococcoides mccartyi species requires acetate as carbon source, but little is known on its growth under acetate limiting conditions. In this study, we observed growth and dechlorination of a D. mccartyi-containing mixed consortium in a fixed-carbon-free medium with trichloroethene in the aqueous phase and H2/CO2 in the headspace. Around 4 mM formate was produced by day 40, while acetate was constantly below 0.05 mM. Microbial community analysis of the consortium revealed dominance by D. mccartyi and Desulfovibrio sp. (57 and 22% 16S rRNA gene copies, respectively). From this consortium, Desulfovibrio sp. strain F1 was isolated and found to produce formate and acetate (1.2 mM and 48 µM, respectively, by day 24) when cultivated alone in the above mentioned medium without trichloroethene. An established co-culture of strain F1 and D. mccartyi strain 195 demonstrated that strain 195 could grow and dechlorinate using acetate produced by strain F1; and that acetate was constantly below 25 µM in the co-culture. To verify that such low level of acetate is utilizable by D. mccartyi, we cultivated strain 195 alone under acetate-limiting conditions and found that strain 195 consumed acetate to below detection (5 µM). Based on the acetate consumption and cell yield of D. mccartyi, we estimated that on average 1.2 × 108 acetate molecules are needed to supply carbon for one D. mccartyi cell. Our study suggests that Desulfovibrio may supply a steady but low amount of fixed carbon to dechlorinating bacteria, exhibiting important implications for natural bio-attenuation when fixed carbon is limited.




Similar content being viewed by others
References
Abelson PH (1990) Inefficient remediation of ground-water pollution. Science 250:733
Aulenta F, Gossett JM, Papini MP, Rossetti S, Majone M (2005) Comparative study of methanol, butyrate, and hydrogen as electron donors for long-term dechlorination of tetrachloroethene in mixed anerobic cultures. Biotechnol Bioeng 91:743–753
Bramono SE, Lam YS, Ong SL, He J (2011) A mesophilic Clostridium species that produces butanol from monosaccharides and hydrogen from polysaccharides. Bioresour Technol 102:9558–9563
Brysch K, Schneider C, Fuchs G, Widdel F (1987) Lithoautotrophic growth of sulfate-reducing bacteria, and description of Desulfobacterium autotrophicum gen. nov., sp. nov. Arch Microbiol 148:264–274
Chen KY, Morris JC (1972) Kinetics of oxidation of aqueous sulfide by oxygen. Environ Sci Technol 6:529–537
Cheng D, Chow WL, He J (2010) A Dehalococcoides-containing co-culture that dechlorinates tetrachloroethene to trans-1,2-dichloroethene. ISME J 4:88–97
Costa C, Teixeira M, Moura I, LeGall J, Moura JJG, Moura I (1997) Formate dehydrogenase from Desulfovibrio desulfuricans ATCC 27774: Isolation and spectroscopic characterization of the active sites (heme, iron-sulfur centers and molybdenum). J Biol Inorg Chem 2:198–208
Ding C, Chow WL, He J (2013) Isolation of Acetobacterium sp. strain AG that reductively debrominates octa- and penta- brominated diphenyl ether technical mixtures. Appl Environ Microbiol 79:1110–1117
Ding C, Zhao S, He J (2014) A Desulfitobacterium sp. strain PR reductively dechlorinates both 1,1,1-trichloroethane and chloroform. Environ Microbiol 16:3387–3397
Dolfing J, Jiang B, Henstra AM, Stams AJM, Plugge CM (2008) Syntrophic growth on formate: a new microbial niche in anoxic environments. Appl Environ Microbiol 74:6126–6131
Drake H, Küsel K, Matthies C (2006) Acetogenic prokaryotes. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The Prokaryotes. Springer, New York, pp 354–420
Fichtel K, Mathes F, Könneke M, Cypionka H, Engelen B (2012) Isolation of sulfate-reducing bacteria from sediments above the deep-subseafloor aquifer. Front Microbiol 3:65
Finneran KT, Forbush HM, VanPraagh CVG, Lovley DR (2002) Desulfitobacterium metallireducens sp. nov., an anaerobic bacterium that couples growth to the reduction of metals and humic acids as well as chlorinated compounds. Int J Syst Evol Microbiol 52:1929–1935
Fite A, Macfarlane GT, Cummings JH, Hopkins MJ, Kong SC, Furrie E, Macfarlane S (2004) Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut 53:523–529
He J, Holmes VF, Lee PK, Alvarez-Cohen L (2007) Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl Environ Microbiol 73:2847–2853
He J, Ritalahti KM, Aiello MR, Löffler FE (2003) Complete detoxification of vinyl chloride by an anaerobic enrichment culture and identification of the reductively dechlorinating population as a Dehalococcoides species. Appl Environ Microbiol 69:996–1003
He J, Sung Y, Dollhopf ME, Fathepure BZ, Tiedje JM, Löffler FE (2002) Acetate versus hydrogen as direct electron donors to stimulate the microbial reductive dechlorination process at chloroethene-contaminated sites. Environ Sci Technol 36:3945–3952
Holliger C, Hahn D, Harmsen H, Ludwig W, Schumacher W, Tindall B, Vazquez F, Weiss N, Zehnder AJB (1998) Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch Microbiol 169:313–321
Holmes VF, He J, Lee PKH, Alvarez-Cohen L (2006) Discrimination of multiple Dehalococcoides strains in a trichloroethene enrichment by quantification of their reductive dehalogenase genes. Appl Environ Microbiol 72:5877–5883
Jetten MSM, Stams AJM, Zehnder AJB (1992) Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol Lett 88:181–198
Kaufhold T, Schmidt M, Cichocka D, Nikolausz M, Nijenhuis I (2013) Dehalogenation of diverse halogenated substrates by a highly enriched Dehalococcoides-containing culture derived from the contaminated mega-site in Bitterfeld. FEMS Microbiol Ecol 83:176–188
Krumholz LR, Sharp R, Fishbain S (1996) A freshwater anaerobe coupling acetate oxidation to tetrachloroethene dehalogenation. Appl Environ Microbiol 62:4108–4113
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Londry KL, Marais DJD (2003) Stable carbon isotope fractionation by sulfate-reducing bacteria. Appl Environ Microbiol 69:2942–2949
Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Müller JA, Fullerton H, Zinder SH, Spormann AM (2013) Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int J Syst Evol Microbiol 63:625–635
Maymó-Gatell X, Chien YT, Gossett JM, Zinder SH (1997) Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568–1571
McInerney MJ, Mackie RI, Bryant MP (1981) Syntrophic association of a butyrate-degrading bacterium and methanosarcina enriched from bovine rumen fluid. Appl Environ Microbiol 41:826–828
Mechalas BJ, Rittenberg SC (1960) Energy coupling in Desulfovibrio Desulfuricans. J Bacteriol 80:501–507
Men Y, Feil H, Verberkmoes NC, Shah MB, Johnson DR, Lee PK, West KA, Zinder SH, Andersen GL, Alvarez-Cohen L (2012) Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analyses. ISME J 6:410–421
Men Y, Yu K, Bælum J, Gao Y, Tremblay J, Prestat E, Stenuit B, Tringe SG, Jansson J, Zhang T, Alvarez-Cohen L (2017) Metagenomic and metatranscriptomic analyses reveal structure and dynamics of a dechlorinating community containing Dehalococcoides mccartyi and corrinoid-providing microorganisms under cobalamin-limited condition. Appl Environ Microbiol. https://doi.org/10.1128/AEM.03508-16
Muyzer G, Dewaal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Muyzer G, Stams AJM (2008) The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol 6:441–454
Müller V (2003) Energy conservation in acetogenic bacteria. Appl Environ Microbiol 69:6345–6353
Peters V, Janssen PH, Conrad R (1999) Transient production of formate during chemolithotrophic growth of anaerobic microorganisms on hydrogen. Curr Microbiol 38:285–289
Rabus R, Hansen TA, Widdel F (2006) Dissimilatory sulfate- and sulfur-reducing prokaryotes. Prokaryotes 2:659–768
Ramsburg CA, Abriola LM, Pennell KD, Löffler FE, Gamache M, Amos BK, Petrovskis EA (2004) Stimulated microbial reductive dechlorination following surfactant treatment at the Bachman Road site. Environ Sci Technol 38:5902–5914
Richardson RE, Bhupathiraju VK, Song DL, Goulet TA, Alvarez-Cohen L (2002) Phylogenetic characterization of microbial communities that reductively dechlorinate TCE based upon a combination of molecular techniques. Environ Sci Technol 36:2652–2662
Ritalahti KM, Löffler FE (2004) Populations implicated in anaerobic reductive dechlorination of 1,2-dichloropropane in highly enriched bacterial communities. Appl Environ Microbiol 70:4088–4095
Schink B (1997) Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61:262–280
Schneidewind U, Haest PJ, Atashgahi S, Maphosa F, Hamonts K, Maesen M, Calderer M, Seuntjens P, Smidt H, Springael D, Dejonghe W (2014) Kinetics of dechlorination by Dehalococcoides mccartyi using different carbon sources. J Contam Hydrol 157:25–36
Scholz-Muramatsu H, Neumann A, Meßmer M, Moore E, Diekert G (1995) Isolation and characterization of Dehalospirillum multivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch Microbiol 163:48–56
Sharma PK, McCarty PL (1996) Isolation and characterization of a facultatively aerobic bacterium that reductively dehalogenates tetrachloroethene to cis-1,2-dichloroethene. Appl Environ Microbiol 62:761–765
Wei N, Finneran KT (2013) Low and high acetate amendments are equally as effective at promoting complete dechlorination of trichloroethylene (TCE). Biodegradation 24:413–425
Acknowledgements
This work was supported by Ng Teng Fong Charitable Foundation (NTFCF) Fund [Grant Number R302-000-198-720] and the National Research Foundation, Prime Minister’s Office, Singapore under the Competitive Research Programme [Grant Number NRF-CRP5-2009-05]. We thank Professor Stephen H. Zinder at Cornell University for providing D. mccartyi strain 195.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ding, C., Alvarez-Cohen, L. & He, J. Growth of Dehalococcoides mccartyi species in an autotrophic consortium producing limited acetate. Biodegradation 29, 487–498 (2018). https://doi.org/10.1007/s10532-018-9846-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-018-9846-9