Abstract
The present research work explores the Nattokinase (NK) producing capacity of five Bacillus subtilis strains (MTCC 2616, MTCC 2756, MTCC 2451, MTCC 1427, and MTCC 7164) using soybean varieties as substrate under solid-state fermentation conditions. Subsequently, the biochemical attributes of NKs were analyzed. Soybean variety didn’t affect the production of NK to a significant extent; however, the five strains differed substantially for their NK producing capacity. NK produced by MTCC 2451 (R3) showed a low Kmvalue implying its higher specificity for fibrin but this strain (MTCC 2451) didn’t produce NK in sufficient quantity. The low Km of MTCC 2451 NK implicates its potential candidature for treating blood clots in cardiovascular patients. The NK produced by MTCC 2616 (R1) was produced in sufficient quantity and showed good fibrin dissolving potential. The aprN of MTCC 2616 substantially varied from the other four strains. The aprN of MTCC 2756 (R2), MTCC 2451 (R3), MTCC 1427 (R4), and MTCC 7164 (R5) shared > 99% sequence identity, but the encoded NKs had significant variations in their Km values. The biochemical-molecular analyses indicate the co-presence of three critical residues (Thr130, Asp140, and Tyr217) as a quintessential attribute in determining the low Km of NK enzymes, and the absence of any one of the three critical residues may affect (highly increase) the Km.




Similar content being viewed by others
References
Adivitiya, Khasa YP (2017) The evolution of recombinant thrombolytics: current status and future directions. Bioengineered 8(4):331–358
Ahmad S, Gromiha MM (2002) NETASA: neural network based prediction of solvent accessibility. Bioinformatics 18(6):819–824
Ahmad S, Gromiha M, Fawareh H, Sarai A (2004) ASAView: database and tool for solvent accessibility representation in proteins. BMC Bioinformatics 5(1):1–5
Akhmetshin RR (1998) Mechanistic investigations of catalysis by some hemostatic and thrombolytic enzymes. The Catholic University of America
Berg JM, Tymoczko JL, Stryer L (2002) Section 8.4 The Michaelis-Menten model accounts for the kinetic properties of many enzymes. In: Freeman WH (ed) Biochemistry. Freeman and Company, New York
Bhargav S, Panda BP, Ali M, Javed S (2008) Solid-state fermentation: an overview. Chem Biochem Eng Q 22(1):49–70
Bhatt PC, Kapoor R, Panda BP (2015) Purification and biochemical characterization of menaquione-7 free, pure nattokinase from Bacillus subtilis. MTCC 2616 52:248–253
Bowie JU, Luthy R, Eisenberg D (1991) A method to identify protein sequences that fold into a known three-dimensional structure. Science 253(5016):164–170
Chandra S, Khowal S, Wajid S (2014) In-silico scrutiny and molecular docking analysis for beta secretase-1 and presenilin-1. Int J Pharma Bio Sci 5(4):274–292
Chang C-T, Fan M-H, Kuo F-C, Sung H-Y (2000) Potent fibrinolytic enzyme from a mutant of Bacillus subtilis IMR-NK1. J Agric Food Chem 48(8):3210–3216
Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ et al (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D 66(1):12–21
Chen H, McGowan EM, Ren N, Lal S, Nassif N, Shad-Kaneez F et al (2018) Nattokinase: a promising alternative in prevention and treatment of cardiovascular diseases. Biomarker Insights 13:1177271918785130
Cho Y-H, Song JY, Kim KM, Kim MK, Lee IY, Kim SB et al (2010) Production of nattokinase by batch and fed-batch culture of Bacillus subtilis. New Biotechnol 27(4):341–346
Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X et al (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35(suppl_2):W375–W383
Deepak V, Kalishwaralal K, Ramkumarpandian S, Babu SV, Senthilkumar S, Sangiliyandi G (2008) Optimization of media composition for Nattokinase production by Bacillus subtilis using response surface methodology. Biores Technol 99(17):8170–8174
Deepak V, Kalishwaralal K, Gurunathan S (2009) Purification, immobilization, and characterization of nattokinase on PHB nanoparticles. Biores Technol 100(24):6644–6646
Diaz AB, Blandino A, Webb C, Caro I (2016) Modelling of different enzyme productions by solid-state fermentation on several agro-industrial residues. Appl Microbiol Biotechnol 100(22):9555–9566
Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res 34(suppl_2):W116–W118
Dym O, Eisenberg D, Yeates T (2012) ERRAT
Fujita M, Ito Y, Hong K, Nishimuro S (1995) Characterization of nattokinase-degraded products from human fibrinogen or cross-linked fibrin. Fibrinolysis 9(3):157–164
Gaziano TA, Bitton A, Anand S, Abrahams-Gessel S, Murphy A (2010) Growing epidemic of coronary heart disease in low- and middle-income countries. Curr Probl Cardiol 35(2):72–115
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 11:681
Gianforcaro A, Kurz M, Guyette FX, Callaway CW, Rittenberger JC, Elmer J (2017) Association of antiplatelet therapy with patient outcomes after out-of-hospital cardiac arrest. Resuscitation 121:98–103
Hatakeyama S, Kafuku M, Okamoto T, Kakizaki Y, Shimasaki N, Fujie N et al (2016) studies on the anticancer mechanisms of the natto extract. J Soc Mater Eng Resour Jpn 27:15–19
Havranek EP, Mujahid MS, Barr DA, Blair IV, Cohen MS, Cruz-Flores S et al (2015) Social determinants of risk and outcomes for cardiovascular disease: a scientific statement from the American Heart Association. Circulation 132(9):873–898
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–38
Huy D, Hao P, Hung P (2016) Screening and identification of Bacillus sp. isolated from traditional Vietnamese soybean-fermented products for high fibrinolytic enzyme production. Int Food Res J 23(1):326
Jensen GS, Lenninger M, Ero MP, Benson KF (2016) Consumption of nattokinase is associated with reduced blood pressure and von Willebrand factor, a cardiovascular risk marker: results from a randomized, double-blind, placebo-controlled, multicenter North American clinical trial. IBPC 9:95
Kapoor R, Panda BP (2013a) Bioprospecting of soybean for production of nattokinase. Nutrafoods 12(3):89–95
Kapoor R, Panda BP (2013b) Production of nattokinase from Bacillus sp. by solid state fermentation. Int J Med Sci Biotechnol 61–65
Kapoor R, Pathak S, Najmi AK, Aeri V, Panda BP (2014) Processing of soy functional food using high pressure homogenization for improved nutritional and therapeutic benefits. Innov Food Sci Emerg Technol 26:490–497
Kapoor R, Harde H, Jain S, Panda AK, Panda BP (2015) Downstream processing, formulation development and antithrombotic evaluation of microbial nattokinase. J Biomed Nanotechnol 11(7):1213–1224
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6):845–858
Khowal S, Siddiqui MZ, Ali S, Khan MT, Khan MA, Naqvi SH et al (2017) A report on extensive lateral genetic reciprocation between arsenic resistant Bacillus subtilis and Bacillus pumilus strains analyzed using RAPD-PCR. Mol Phylogenet Evol 107:443–454
Khowal S, Naqvi SH, Monga S, Jain SK, Wajid S (2018a) Assessment of cellular and serum proteome from tongue squamous cell carcinoma patient lacking addictive proclivities for tobacco, betel nut, and alcohol: case study. J Cell Biochem 119(7):5186–5221
Khowal S, Jain SK, Wajid S (2018b) A potential association between mutations in the iNOS cDNA 3′ stretch and oral squamous cell carcinoma-A preliminary study. Meta Gene 16:189–195
Khowal S, Monga S, Naqvi SH, Jain SK, Wajid S (2021) Molecular winnowing, expressional analyses and interactome scrutiny of cellular proteomes of oral squamous cell carcinoma. Adv Cancer Biol 2:100003
Kou Y, Feng R, Chen J, Duan L, Wang S, Hu Y et al (2020) Development of a nattokinase–polysialic acid complex for advanced tumor treatment. Eur J Pharm Sci 145:105241
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870
Kumar V, Ahluwalia V, Saran S, Kumar J, Patel AK, Singhania RR (2021) Recent developments on solid-state fermentation for production of microbial secondary metabolites: challenges and solutions. Bioresour Technol 323:124566
Kurosawa Y, Nirengi S, Homma T, Esaki K, Ohta M, Clark JF et al (2015) A single-dose of oral nattokinase potentiates thrombolysis and anti-coagulation profiles. Sci Rep 5(1):11601
Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S et al (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43(W1):W580–W584
Liu J, Xing J, Chang T, Ma Z, Liu H (2005) Optimization of nutritional conditions for nattokinase production by Bacillus natto NLSSE using statistical experimental methods. Process Biochem 40(8):2757–2762
Mahajan PM, Gokhale SV, Lele SS (2010) Production of nattokinase using Bacillus natto NRRL 3666: media optimization, scale up, and kinetic modeling. Food Sci Biotechnol 19(6):1593–1603
Mbah AN, Isokpehi RD (2013) Identification of functional regulatory residues of the β-lactam inducible penicillin binding protein in methicillin-resistant Staphylococcus aureus. Chemother Res Pract 2013:614670
McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N et al (2013) Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41(W1):W597–W600
Milner M (2008) Nattokinase: a potent and safe thrombolytic. FOCUS Newsletter 2
Milner M, Makise K (2002) Natto and its active ingredient nattokinase: a potent and safe thrombolytic agent. Altern Complement Ther 8(3):157–164
Nakamura T, Yamagata Y, Ichishima E (1992) Nucleotide sequence of the subtilisin NAT gene, aprN, of Bacillus subtilis (natto). Biosci Biotechnol Biochem 56(11):1869–1871
Nguyen TT, Quyen TD, Le HT (2013) Cloning and enhancing production of a detergent- and organic-solvent-resistant nattokinase from Bacillus subtilis VTCC-DVN-12-01 by using an eight-protease-gene-deficient Bacillus subtilis WB800. Microb Cell Fact. https://doi.org/10.1186/1475-2859-12-79
Noori HR, Spanagel R (2013) In silico pharmacology: drug design and discovery’s gate to the future. In Silico Pharmacol. https://doi.org/10.1186/2193-9616-1-1
Oriol E, Raimbault M, Roussos S, Viniegra-Gonzales G (1988) Water and water activity in the solid state fermentation of cassava starch by Aspergillus niger. Appl Microbiol Biotechnol 27(5):498–503
Prajapat R, Jain S, Vaishnav MK, Sogani S (2020) Structural modeling and validation of growth/differentiation factor 15 [NP_004855] associated with pregnancy complication-hyperemesis gravidarum. J Krishna Inst Med Sci (JKIMSU) 9(3)
Robinson PK (2015) Enzymes: principles and biotechnological applications. Essays Biochem 59:1–41
Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM et al (2020) Global burden of cardiovascular diseases and risk factors, 1990–2019: update from the GBD 2019 study. J Am Coll Cardiol 76(25):2982–3021
Sanbrook J, Fritsch E, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor
Schrödinger L (2016) The PyMOL molecular graphics system, Version 1.8 Schrödinger, LLC
Seibert E, Tracy TS (2014) Fundamentals of enzyme kinetics. In: Enzyme kinetics in drug metabolism, pp 9–22
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7(1):539
Sippl MJ (1993) Recognition of errors in three-dimensional structures of proteins. Proteins 17(4):355–362
Soares VF, Castilho LR, Bon EP, Freire DM (2005) High-yield Bacillus subtilis protease production by solid-state fermentation. Appl Biochem Biotechnol 121–124:311–319
Soccol CR, Costa ESF, Letti LAJ, Karp SG, Woiciechowski AL, Vandenberghe LPS (2017) Recent developments and innovations in solid state fermentation. Biotechnol Res Innov 1(1):52–71
Stark K, Massberg S (2021) Interplay between inflammation and thrombosis in cardiovascular pathology. Nat Rev Cardiol 18:666–682
Sumi H, Hamada H, Tsushima H, Mihara H, Muraki H (1987) A novel fibrinolytic enzyme (nattokinase) in the vegetable cheese Natto; a typical and popular soybean food in the Japanese diet. Experientia 43(10):1110–1111
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10(3):512–526
Undas A, Ariëns RAS (2011) Fibrin clot structure and function. Arterioscler Thromb Vasc Biol 31(12):e88–e99
Urano T, Ihara H, Umemura K, Suzuki Y, Oike M, Akita S et al (2001) The profibrinolytic enzyme subtilisin NAT purified from Bacillus subtilis cleaves and inactivates plasminogen activator inhibitor Type 1. J Biol Chem 276(27):24690–24696
Versteeg HH, Heemskerk JWM, Levi M, Reitsma PH (2013) New fundamentals in hemostasis. Physiol Rev 93(1):327–358
Wang S-L, Yeh P-Y (2006) Production of a surfactant-and solvent-stable alkaliphilic protease by bioconversion of shrimp shell wastes fermented by Bacillus subtilis TKU007. Process Biochem 41(7):1545–1552
Wang C, Du M, Zheng D, Kong F, Zu G, Feng Y (2009) Purification and characterization of nattokinase from Bacillus subtilis natto B-12. J Agric Food Chem 57(20):9722–9729
Wei X, Luo M, Xie Y, Yang L, Li H, Xu L et al (2012) Strain screening, fermentation, separation, and encapsulation for production of nattokinase functional food. Appl Biochem Biotechnol 168(7):1753–1764
Weng M, Deng X, Bao W, Zhu L, Wu J, Cai Y et al (2015) Improving the activity of the subtilisin nattokinase by site-directed mutagenesis and molecular dynamics simulation. Biochem Biophys Res Commun 465(3):580–586
Weng Y, Yao J, Sparks S, Wang KY (2017) Nattokinase: an oral antithrombotic agent for the prevention of cardiovascular disease. Int J Mol Sci 18(3):523
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35(suppl_2):W407–W410
Wilson K (2001) Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol 56(1):2.4.1-2.4.5
Wolberg AS (2016) Primed to understand fibrinogen in cardiovascular disease. Arterioscler Thromb Vasc Biol 36(1):4–6
Wright ES, Yilmaz LS, Noguera DR (2012) DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol 78(3):717–725
Wu S, Feng C, Zhong J, Huan L (2007) Roles of s3 site residues of nattokinase on its activity and substrate specificity. J Biochem 142(3):357–364
Yanagisawa Y, Chatake T, Chiba-Kamoshida K, Naito S, Ohsugi T, Sumi H et al (2010) Purification, crystallization and preliminary X-ray diffraction experiment of nattokinase from Bacillus subtilis natto. Acta Crystallogr F 66(12):1670–1673
Yanagisawa Y, Chatake T, Naito S, Ohsugi T, Yatagai C, Sumi H et al (2013) X-ray structure determination and deuteration of nattokinase. J Synchrotron Radiat 20(6):875–879
Yin L-J, Lin H-H, Jiang S-T (2010) Bioproperties of potent nattokinase from Bacillus subtilis YJ1. J Agric Food Chem 58(9):5737–5742
Yongmin Y et al (2019) Nattokinase crude extract inhibits hepatocellular carcinoma growth in mice. J Microbiol Biotechnol 29(8):1281–1287
You C, Zhang C, Kong F, Feng C, Wang J (2016) Comparison of the effects of biocontrol agent Bacillus subtilis and fungicide metalaxyl–mancozeb on bacterial communities in tobacco rhizospheric soil. Ecol Eng 91:119–125
Zheng Z, Zuo Z, Liu Z, Tsai K, Liu A, Zou G (2005) Construction of a 3D model of nattokinase, a novel fibrinolytic enzyme from Bacillus natto A novel nucleophilic catalytic mechanism for nattokinase. J Mol Graph Model 23(4):373–380
Zheng Z, Ye M, Zuo Z, Liu Z, Tai K, Zou G (2006) Probing the importance of hydrogen bonds in the active site of the subtilisin nattokinase by site-directed mutagenesis and molecular dynamics simulation. Biochem J 395(3):509–515
Acknowledgements
The authors acknowledge the Department of Science and Technology, Govt. of India for providing fellowship to research scholar under the INSPIRE programme.
Author information
Authors and Affiliations
Contributions
RK performed fermentation experiments, analyzed the data, conducted manuscript writing, and manuscript finalization. SK performed molecular experiments, in-silico analyses, analyzed the data, conducted manuscript writing, and manuscript finalization. BPP and SW conceived the idea, analyzed the data and reviewed the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors report no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
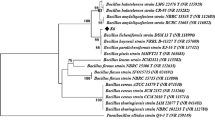
The aprN sequences of the five bacterial strains have been submitted to GenBank with accession numbers as KJ205599 for R1 (B. subtilis MTCC 2616); KJ205600 for R2 (B. subtilis MTCC 2756); KJ144819 for R3 (B. subtilis MTCC 2451), KJ174339 for R4 (B. subtilis MTCC 1427) and KJ174338 for R5 (B. subtilis MTCC 7164).
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kapoor, R., Khowal, S., Panda, B.P. et al. Comparative genomic analyses of Bacillus subtilis strains to study the biochemical and molecular attributes of nattokinases. Biotechnol Lett 44, 485–502 (2022). https://doi.org/10.1007/s10529-022-03226-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-022-03226-1