Abstract
Objectives
To discover novel feruloyl esterases (FAEs) by the function-driven screening procedure from soil metagenome.
Results
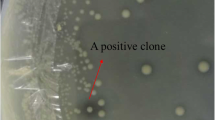
A novel FAE gene bds4 was isolated from a soil metagenomic library and over-expressed in Escherichia coli. The recombinant enzyme BDS4 was purified to homogeneity with a predicted molecular weight of 38.8 kDa. BDS4 exhibited strong activity (57.05 U/mg) toward methyl ferulate under the optimum pH and temperature of 8.0 and 37°C. Based on its amino acid sequence and model substrates specificity, BDS4 was classified as a type-C FAE. The quantity of the releasing ferulic acid can be enhanced significantly in the presence of xylanase compared with BDS4 alone from de-starched wheat bran. In addition, BDS4 can also hydrolyze several phthalates such as diethyl phthalate, dimethyl phthalate and dibutyl phthalate.
Conclusion
The current investigation discovered a novel FAE with phthalate-degrading activity and highlighted the usefulness of metagenomic approaches as a powerful tool for discovery of novel FAEs.







Similar content being viewed by others
References
Altschul SF, Wootton JC, Gertz EM et al (2005) Protein database searches using compositionally adjusted substitution matrices. FEBS J 272(20):5101–5109
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Benjamin S, Pradeep S, Sarath Josh M et al (2015) A monograph on the remediation of hazardous phthalates. J Hazard Mater 298:58–72
Benoit I, Danchin EGJ, Bleichrodt RJ, De Vries RP (2008) Biotechnological applications and potential of fungal feruloyl esterases based on prevalence, classification and biochemical diversity. Biotech Lett 30:387–396. https://doi.org/10.1007/s10529-007-9564-6
Brady SF (2007) Construction of soil environmental DNA cosmid libraries and screening for clones that produce biologically active small molecules. Nat Protoc 2:1297–1305
Chen X, Zhang X, Yang Y et al (2015) Biodegradation of an endocrine-disrupting chemical di-n-butyl phthalate by newly isolated Camelimonas sp. and enzymatic properties of its hydrolase. Biodegradation 26:171–182
Cheng F, Sheng J, Cai T et al (2012a) A protease-insensitive feruloyl esterase from China Holstein cow rumen metagenomic library: expression, characterization, and utilization in ferulic acid release from wheat straw. J Agric Food Chem 60:2546–2553. https://doi.org/10.1021/jf204556u
Cheng F, Sheng J, Dong R et al (2012b) Novel xylanase from a holstein cattle rumen metagenomic library and its application in xylooligosaccharide and ferulic Acid production from wheat straw. J Agric Food Chem 60:12516–12524
Crepin VF, Faulds CB, Connerton IF (2004) Functional classification of the microbial feruloyl esterases. Appl Microbiol Biotechnol 63:647–652. https://doi.org/10.1007/s00253-003-1476-3
Dilokpimol A, Mäkelä MR, Aguilar-Pontes MV et al (2016) Diversity of fungal feruloyl esterases: updated phylogenetic classification, properties, and industrial applications. Biotechnol Biofuels 9:1–18. https://doi.org/10.1186/s13068-016-0651-6
Dilokpimol A, Mäkelä MR, Varriale S et al (2018) Fungal feruloyl esterases: functional validation of genome mining based enzyme discovery including uncharacterized subfamilies. New Biotechnol 41:9–14. https://doi.org/10.1016/j.nbt.2017.11.004
Gopalan N, Rodríguez-Duran LV, Saucedo-Castaneda G, Nampoothiri KM (2015) Review on technological and scientific aspects of feruloyl esterases: a versatile enzyme for biorefining of biomass. Biores Technol 193:534–544. https://doi.org/10.1016/j.biortech.2015.06.117
Haase-Aschoff P, Linke D, Nimtz M et al (2013) An enzyme from Auricularia auricula-judae combining both benzoyl and cinnamoyl esterase activity. Process Biochem 48:1872–1878
Hancock JM, Bishop MJ (2004) Dictionary of bioinformatics and computational biology. ORF Finder (Open Reading Frame Finder). Wiley, New Jersey
Handelsman J (2004) Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68:669–685
Koseki T, Hori A, Seki S et al (2009) Characterization of two distinct feruloyl esterases, AoFaeB and AoFaeC, from Aspergillus oryzae. Appl Microbiol Biotechnol 83:689–696. https://doi.org/10.1007/s00253-009-1913-z
Li W, Cowley A, Uludag M et al (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43:W580–W584. https://doi.org/10.1093/nar/gkv279
Li X, Guo J, Hu Y et al (2018) Identification of a novel feruloyl esterase by functional screening of a soil metagenomic library. Appl Biochem Biotechnol 187(1):1–14
Lu Y, Tang F, Wang Y et al (2009) Biodegradation of dimethyl phthalate, diethyl phthalate and di-n-butyl phthalate by Rhodococcus sp. L4 isolated from activated sludge. J Hazard Mater 168:938–943
MacOn MB, Fenton SE (2013) Endocrine disruptors and the breast: early life effects and later life disease. J Mammary Gland Biol Neoplasia 18(1):43–61
Moukouli M, Topakas E, Christakopoulos P (2008) Cloning, characterization and functional expression of an alkalitolerant type C feruloyl esterase from Fusarium oxysporum. Appl Microbiol Biotechnol 79:245–254. https://doi.org/10.1007/s00253-008-1432-3
Ohlhoff CW, Kirby BM, Van Zyl L et al (2015) An unusual feruloyl esterase belonging to family VIII esterases and displaying a broad substrate range. J Mol Catal B Enzym 118:79–88
Oliveira DM, Mota TR, Oliva B et al (2019) Feruloyl esterases: biocatalysts to overcome biomass recalcitrance and for the production of bioactive compounds. Biores Technol 278:408–423. https://doi.org/10.1016/j.biortech.2019.01.064
Pereira MR, Maester TC, Mercaldi GF et al (2017) From a metagenomic source to a high-resolution structure of a novel alkaline esterase. Appl Microbiol Biotechnol 101:4935–4949. https://doi.org/10.1007/s00253-017-8226-4
Prates JA, Tarbouriech N, Charnock SJ et al (2001) The structure of the feruloyl esterase module of xylanase 10B from Clostridium thermocellum provides insights into substrate recognition. Structure 9(12):1183–1190
Record E, Asther M, Sigoillot C et al (2003) Overproduction of the Aspergillus niger feruloyl esterase for pulp bleaching application. Appl Microbiol Biotechnol 62(4):349–355
Reyes-Duarte D, Ferrer M, García-Arellano H (2012) Functional-based screening methods for lipases, esterases, and phospholipases in metagenomic libraries. Methods Mol Biol 861:101
Schloss PD, Handelsman J (2005) Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol 6:1–4
Shahidi F, Chandrasekara A (2010) Hydroxycinnamates and their in vitro and in vivo antioxidant activities. Phytochem Rev 9:147–170. https://doi.org/10.1007/s11101-009-9142-8
Shu LS, Gang L, Xiao PH, Yu HL (2011) Molecular cloning, overexpression and characterization of a novel feruloyl esterase from a soil metagenomic library. J Mol Microbiol Biotechnol 20:196–203
Suzuki K, Hori A, Kawamoto K et al (2014) Crystal structure of a feruloyl esterase belonging to the tannase family: a disulfide bond near a catalytic triad. Proteins: structure. Funct Bioinform 82:2857–2867. https://doi.org/10.1002/prot.24649
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Torsvik V, Goksøyr J, Daae FL (1990) High diversity in DNA of soil bacteria. Appl Environ Microbiol 56:782–787
Torsvik V, Daae FL, Sandaa RA, Ovreås L (1998) Novel techniques for analysing microbial diversity in natural and perturbed environments. J Biotechnol 64:53–62
Wang L, Li Z, Zhu M et al (2016) An acidic feruloyl esterase from the mushroom Lactarius hatsudake: a potential animal feed supplement. Int J Biol Macromol 93:290–295
Williamson G, Kroon PA, Faulds CB (1998) Hairy plant polysaccharides: a close shave with microbial esterases. Microbiology 144:2011–2023
Wong DWS, Chan VJ, Liao H, Zidwick MJ (2013) Cloning of a novel feruloyl esterase gene from rumen microbial metagenome and enzyme characterization in synergism with endoxylanases. J Ind Microbiol Biotechnol 40:287–295. https://doi.org/10.1007/s10295-013-1234-1
Wu M, Abokitse K, Grosse S et al (2012) New feruloyl esterases to access phenolic acids from grass biomass. Appl Biochem Biotechnol 168(1):129–143
Yang J, Roy A, Zhang Y (2013a) BioLiP: a semi-manually curated database for biologically relevant ligand–protein interactions. Nucleic Acids Res 41:1096–1103
Yang J, Roy A, Zhang Y (2013b) Protein–ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 29:2588–2595
Yao J, Chen QL, Shen AX et al (2013) A novel feruloyl esterase from a soil metagenomic library with tannase activity. J Mol Catal B Enzym 95:55–61. https://doi.org/10.1016/j.molcatb.2013.05.026
Yu P, McKinnon JJ, Christensen DA (2011) Hydroxycinnamic acids and ferulic acid esterase in relation to biodegradation of complex plant cell walls. Can J Anim Sci. 85(3):255–267
Acknowledgements
This research was supported by the Fundamental Research Funds for the Central Universities (KYYJ201708) and Qing Lan Project of Jiangsu Province.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Accession Number
The bds4 gene was deposited in the GenBank database with the accession no. MH445495.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, S., Nan, F., Jiang, J. et al. Molecular cloning, expression and characterization of a novel feruloyl esterase from a soil metagenomic library with phthalate-degrading activity. Biotechnol Lett 41, 995–1006 (2019). https://doi.org/10.1007/s10529-019-02693-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-019-02693-3