Abstract
Objectives
This study reports the identification of a new bacterial azoreductase (AzoR) from Klebsiella oxytoca GS-4-08, its heterologous production in Escherichia coli and the decolorization of azo dyes.
Results
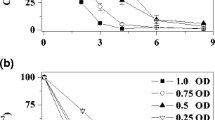
The AzoR, as a flavin-free, oxygen-insensitive enzyme, has a molecular mass of 22 kDa and a high substrate specificity for methyl red (MR). In the presence of 5 mM of NADH, the enzyme activity for decolorization of 50 mg l−1 of MR reached 5.61 μmol min−1 mg protein−1. Double-reciprocal plots indicated that the NADH and MR reductions proceed by ping-pong mechanism. The calculated Vm was 8.17 μM min−1 mg protein−1, and the AzoR retained over 75% of relative activity under temperature range from 20 to 50 °C.
Conclusion
This study for the first time identified an AzoR from Klebsiella oxytoca strain, which enabled efficient degradation of azo dyes by strain GS-4-08.



Similar content being viewed by others
References
Blumel S, Stolz A (2003) Cloning and characterization of the gene coding for the aerobic azoreductase from Pigmentiphaga kullae K24. Appl Microbiol Biotechnol 62:186–190
Burger S, Stolz A (2010) Characterisation of the flavin-free oxygen-tolerant azoreductase from Xenophilus azovorans KF46F in comparison to flavin-containing azoreductases. Appl Microbiol Biotechnol 87:2067–2076
Hu H, Li L, Ding S (2015) An organic solvent-tolerant phenolic acid decarboxylase from Bacillus licheniformis for the efficient bioconversion of hydroxycinnamic acids to vinyl phenol derivatives. Appl Microbiol Biotechnol 99:5071–5081
Khalid A, Arshad M, Crowley DE (2009) Biodegradation potential of pure and mixed bacterial cultures for removal of 4-nitroaniline from textile dye wastewater. Water Res 43:1110–1116
Lang W, Sirisansaneeyakul S, Ngiwsara L, Mendes S, Martins LO, Okuyama M, Kimura A (2013) Characterization of a new oxygen-insensitive azoreductase from Brevibacillus laterosporus TISTR1911: toward dye decolorization using a packed-bed metal affinity reactor. Bioresour Technol 150:298–306
Liu G, Zhou J, Lv H, Xiang X, Wang J, Zhou M, Qv Y (2007) Azoreductase from Rhodobacter sphaeroides AS1.1737 is a flavodoxin that also functions as nitroreductase and flavin mononucleotide reductase. Appl Microbiol Biotechnol 76:1271–1279
Liu WX, Wang QL, Hou JY, Tu C, Luo YM, Christie P (2016) Whole genome analysis of halotolerant and alkalotolerant plant growth-promoting rhizobacterium Klebsiella sp D5A. Sci Rep 6:26710
Matsumoto K, Mukai Y, Ogata D, Shozui F, Nduko JM, Taguchi S, Ooi T (2010) Characterization of thermostable FMN-dependent NADH azoreductase from the moderate thermophile Geobacillus stearothermophilus. Appl Microbiol Biotechnol 86:1431–1438
Mendes S, Pereira L, Batista C, Martins LO (2011) Molecular determinants of azo reduction activity in the strain Pseudomonas putida MET94. Appl Microbiol Biotechnol 92:393–405
Misal SA, Lingojwar DP, Shinde RM, Gawai KR (2011) Purification and characterization of azoreductase from alkaliphilic strain Bacillus badius. Process Biochem 46:1264–1269
Misal SA, Lingojwar DP, Lokhande MN, Lokhande PD, Gawai KR (2014) Enzymatic transformation of nitro-aromatic compounds by a flavin-free NADH azoreductase from Lysinibacillus sphaericus. Biotechnol Lett 36:127–131
Moutaouakkil A, Zeroual Y, Zohra Dzayri F, Talbi M, Lee K, Blaghen M (2003) Purification and partial characterization of azoreductase from Enterobacter agglomerans. Arch Biochem Biophys 413:139–146
Ooi T, Shibata T, Sato R, Ohno H, Kinoshita S, Thuoc TL, Taguchi S (2007) An azoreductase, aerobic NADH-dependent flavoprotein discovered from Bacillus sp.: functional expression and enzymatic characterization. Appl Microbiol Biotechnol 75:377–386
Punj S, John GH (2009) Purification and identification of an FMN-dependent NAD(P)H azoreductase from Enterococcus faecalis. Curr Issues Mol Biol 11:59–65
Qi J, Paul CE, Hollmann F, Tischler D (2017) Changing the electron donor improves azoreductase dye degrading activity at neutral pH. Enzyme Microb Technol 100:17–19
Ramalho PA, Cardoso AH, Cavaco-Paulo A, Ramalho AT (2004) Characterization of azo reduction activity in a novel ascomycete yeast strain. Appl Environ Microb 70:2279–2288
Suzuki Y, Yoda T, Ruhul A, Sugiura W (2001) Molecular cloning and characterization of the gene coding for azoreductase from Bacillus sp OY1-2 isolated from soil. J Biol Chem 276:9059–9065
Suzuki H, Abe T, Doi K, Ohshima T (2018) Azoreductase from alkaliphilic Bacillus sp AO1 catalyzes indigo reduction. Appl Microbiol Biotechnol 102:9171–9181
Verma K, Saha G, Kundu LM, Dubey VK (2019) Biochemical characterization of a stable azoreductase enzyme from Chromobacterium violaceum: application in industrial effluent dye degradation. Int J Biol Macromol 121:1011–1018
Yu L, Li WW, Lam MHW, Yu HQ, Wu C (2012) Isolation and characterization of a Klebsiella oxytoca strain for simultaneous azo-dye anaerobic reduction and bio-hydrogen production. Appl Microbiol Biotechnol 95:255–262
Yu L, Zhang XY, Xie T, Hu JM, Wang S, Li WW (2015) Intracellular azo decolorization is coupled with aerobic respiration by a Klebsiella oxytoca strain. Appl Microbiol Biotechnol 99:2431–2439
Yu L, Cao MY, Wang PT, Wang S, Yue YR, Yuan WD, Qiao WC, Wang F, Song X (2017) Simultaneous decolorization and biohydrogen production from xylose by Klebsiella oxytoca GS-4-08 in the presence of azo dyes with sulfonate and carboxyl groups. Appl Environ Microb 83:e00508–e00517
Zhang X, Ng IS, Chang J-S (2016) Cloning and characterization of a robust recombinant azoreductase from Shewanella xiamenensis BC01. J Taiwan Inst Chem Eng 61:97–105
Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 51778298) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The Authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hua, Jq., Yu, L. Cloning and characterization of a Flavin-free oxygen-insensitive azoreductase from Klebsiella oxytoca GS-4-08. Biotechnol Lett 41, 371–378 (2019). https://doi.org/10.1007/s10529-019-02647-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-019-02647-9