Abstract
Objectives
To test the applicability of Cpf1 from Francisella novivida in genomic integration of in vivo assembled DNA parts in Saccharomyces cerevisiae.
Results
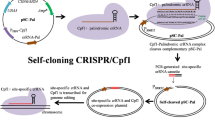
An easy-to-use vector toolkit, containing a CEN6/ARS4 plasmid expressing Cpf1 from Francisella novivida (FnCpf1) and a 2 μ plasmid for crRNA or crRNA array expressing, was constructed for Cpf1-assisted genomic integration in S. cerevisiae. Our results showed that FnCpf1 allowed for targeted singleplex, doubleplex, and tripleplex genomic integration of in vivo assembled DNA parts with efficiencies of 95, 52, and 43%, respectively.
Conclusions
CRISPR-Cpf1 system allows for efficient genomic integration of in vivo assembled DNA parts in S. cerevisiae, and thus provides an alternative CRISPR-Cas method for metabolic pathway engineering in addition to CRISPR-Cas9 system previously reported for yeast.





Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14(2):115–132
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278
Hu H et al (2016) Screening and identification of proteins interacting with IL-24 by the yeast two-hybrid screen, Co-IP, and FRET assays. Anticancer Drugs 27:318
Jakočiūnas T et al (2015) CasEMBLR: Cas9-facilitated multiloci genomic integration of in vivo assembled DNA parts in Saccharomyces cerevisiae. ACS Synth Biol 4:1226
Meadows AL et al (2016) Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature 537:694
Nielsen J, Keasling JD (2016) Engineering cellular metabolism. Cell 164:1185–1197
Ro DK et al (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440:940
Siddiqui MS, Choksi A, Smolke CD (2015) A system for multi-locus chromosomal integration and transformation-free selection marker rescue. FEMS Yeast Res 14:1171–1185
Storici F, Durham CL, Gordenin DA, Resnick MA (2003) Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc Natl Acad Sci USA 100:14994–14999
Swiat MA, Dashko S, Den RM, Wijsman M, van der Oost J, Daran JM, Daran-Lapujade P (2017) FnCpf1: a novel and efficient genome editing tool for Saccharomyces cerevisiae. Nucleic Acids Res 45:12585–12598
Verwaal R, Buiting-Wiessenhaan N, Dalhuijsen S, Roubos JA (2018) CRISPR/Cpf1 enables fast and simple genome editing of Saccharomyces cerevisiae. Yeast 35:201–211. https://doi.org/10.1002/yea.3278
Xie W, Liu M, Lv X, Lu W, Gu J, Yu H (2013) Construction of a controllable β-carotene biosynthetic pathway by decentralized assembly strategy in Saccharomyces cerevisiae. Biotechnol Bioeng 111:125–133
Xie W, Ye L, Lv X, Xu H, Yu H (2015) Sequential control of biosynthetic pathways for balanced utilization of metabolic intermediates in Saccharomyces cerevisiae. Metab Eng 28:8
Zetsche et al (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759
Zetsche B et al (2017) Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat Biotechnol 35:31
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (No. 31500043). The authors sincerely thank Prof. Hongwei Yu (Zhejiang University) for providing plasmids of pESC-LEU-cen, pESC-URA, pMRI-34-crtE-tHMG1, pMRI-35-crtYB-crtI, pUMRI-15, and strain of S. cerevisiae BY4741.
Supporting information
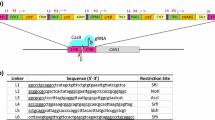
Supplementary Fig. 1—Composition of the CRISPR arrays for singleplex, doubleplex, and tripleplex genome editing target sites.
Supplementary Fig. 2—Graphical depiction of the positions of the genomic target sites of the Gal1-7, Gal80, and HO for FnCpf1.
Supplementary Fig. 3—Pictures of transformants to determine the efficiency of targeted singleplex, doubleplex, and tripleplex genomic integration facilitated by FnCpf1. A: Pictures of transformants obtained by co-transforming donor DNA of MEL1 with plasmid expressing singleplex crRNA (Gal1-7_pos1, Gal1-7_pos2, and Gal1-7_pos3) or without crRNA plasmid (control1). B: Pictures of transformants obtained by co-transforming donor DNA of MEL1 and KanMX with plasmid expressing doubleplex crRNA array (Gal1-7_pos2 + Gal80_pos1, Gal1-7_pos2 + Gal80_pos2, and Gal1-7_pos2 + Gal-80_pos3) or without crRNA plasmid (control2). C: Pictures of transformants obtained by co-transforming donor DNA of crtI, crtYB, and tHMG1-crtE with plasmid expressing tripleplex crRNA array (Gal1-7_pos2 + Gal80_pos1 + HO_pos1, Gal1-7_pos2 + Gal80_pos1 + HO_pos2, and Gal1-7_pos2 + Gal80_pos1 + HO_pos3) or without crRNA plasmid (control3).
Supplementary Table 1—Primers and oligonucleotides used in this study.
Supplementary Table 2—Donor DNA flank and expression cassette sequences.
Supplementary Table 3—Synthesized DNA fragments in this study.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, ZH., Liu, M., Wang, FQ. et al. Cpf1-assisted efficient genomic integration of in vivo assembled DNA parts in Saccharomyces cerevisiae. Biotechnol Lett 40, 1253–1261 (2018). https://doi.org/10.1007/s10529-018-2574-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-018-2574-8