Abstract
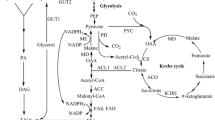
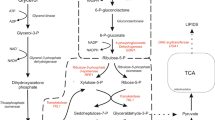
Malic enzyme (EC 1.1.1.40) converts l-malate to pyruvate and CO2 providing NADPH for metabolism especially for lipid biosynthesis in oleaginous microorganisms. However, its role in the oleaginous yeast, Yarrowia lipolytica, is unclear. We have cloned the malic enzyme gene (YALI0E18634g) from Y. lipolytica into pET28a, expressed it in Escherichia coli and purified the recombinant protein (YlME). YlME used NAD+ as the primary cofactor. Km values for NAD+ and NADP+ were 0.63 and 3.9 mM, respectively. Citrate, isocitrate and α-ketoglutaric acid (>5 mM) were inhibitory while succinate (5–15 mM) increased NADP+- but not NAD+-dependent activity. To determine if fatty acid biosynthesis could be increased in Y. lipolytica by providing additional NADPH from an NADP+-dependent malic enzyme, the malic enzyme gene (mce2) from an oleaginous fungus, Mortierella alpina, was expressed in Y. lipolytica. No significant changes occurred in lipid content or fatty acid profiles suggesting that malic enzyme is not the main source of NADPH for lipid accumulation in Y. lipolytica.





Similar content being viewed by others
References
Ageitos JM, Vallejo JA, Veiga-Crespo P, Villa TG (2011) Oily yeasts as oleaginous cell factories. Appl Microbiol Biotechnol 90:1219–1225
Athenstaedt K (2011) YALI0E32769g (DGA1) and YALI0E16797g (LRO1) encode major triacylglycerol synthases of the oleaginous yeast Yarrowia lipolytica. Biochim Biophys Acta 1811:587–596
Barth G, Gaillardin C (1996) Non-conventional yeasts in biotechnology. Springer, Berlin
Beopoulos A, Mrozova Z, Thevenieau F, Le Dall MT, Hapala I, Papanikolaou S, Chardot T, Nicaud JM (2008) Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl Environ Microbiol 74:7779–7789
Beopoulos A, Chardot T, Nicaud JM (2009) Yarrowia lipolytica: a model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie 91:692–696
Beopoulos A, Desfougéres T, Sabirova J, Nicaud JM (2010) Yarrowia lipolytica as a cell factory for oleochemical biotechnology. In: Timmis KN (ed) Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, pp 3003–3010
Beopoulos A, Nicaud JM, Gaillardin C (2011) An overview of lipid metabolism in yeasts and its impact on biotechnological processes. Appl Microbiol Biotechnol 90:1193–1206
Blank LM, Lehmbeck F, Sauer U (2005) Metabolic-flux and network analysis in fourteen hemiascomycetous yeasts. FEMS Yeast Res 5:545–558
Blazeck J, Liu L, Redden H, Alper H (2011) Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl Environ Microbiol 77:7905–7914
Dujon B, Sherman D, Fischer G et al (2004) Genome evolution in yeasts. Nature 430:35–44
Evans CT, Ratledge C (1984) Effect of nitrogen source on lipid accumulation in oleaginous yeasts. Microbiology 30:1693–1704
Evans CT, Ratledge C (1985) The physiological significance of citric acid in the control of metabolism in lipid-accumulating yeasts. Biotechnol Genet Eng Rev 3:349–375
Hsu RY, Lardy HA (1969) Malic enzyme. Methods Enzymol 13:230–235
Kendrick A, Ratledge C (1992) Lipid formation in the oleaginous mould Entomophthora exitalis grown in continuous culture: effects of growth rate, temperature and dissolved oxygen tension on polyunsaturated fatty acids. Appl Microbiol Biotechnol 3:18–22
Li Z, Sun H, Mo X, Li X, Xu B, Tian P (2013) Overexpression of malic enzyme (ME) of M. circinelloides improved lipid accumulation in engineered Rhodotorula glutinis. Appl Microbiol Biotechnol 97:4927–4936
Liu H, Zhao X, Wang F, Li Y, Jiang X, Ye M, Zhao ZK, Zou H (2009) Comparative proteomic analysis of Rhodosporidium toruloides during lipid accumulation. Yeast 26:553–566
Liu H, Zhao X, Jiang X, Wang F, Zhang S, Ye M, Zhao ZK, Zou H (2011) The proteome analysis of oleaginous yeast Lipomyces starkeyi. FEMS Yeast Res 11:42–51
Morin N, Cescut J, Beopoulos A, Lelandais G, Le Berre V, Uribelarrea JL, Molina-Jouve C, Nicaud JM (2011) Transcriptomic analyses during the transition from biomass production to lipid accumulation in the oleaginous yeast Yarrowia lipolytica. PLoS ONE 6:e27966
Papanikolaou S, Chatzifragkou A, Fakas S, Panayotou MG, Komaitis M, Nicaud JM, Aggelis G (2009) Biosynthesis of lipids and organic acids by Yarrowia lipolytica strains cultivated on glucose. Eur J Lipid Sci Technol 111:1221–1232
Pon J, Napoli E, Luckhart S, Giulivi C (2011) Mitochondrial NAD+-dependent malic enzyme from Anopheles stephensi: a possible novel target for malaria mosquito control. Malar J 26(10):318. doi:10.1186/1475-2875-10-318
Ratledge C, Wynn JP (2002) The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv Appl Microbiol 51:1–51
Su KL, Chang KY, Hung HC (2009) Effects of structural analogues of the substrate and allosteric regulator of the human mitochondrial NAD(P)+-dependent malic enzyme. Bioorg Med Chem 17:5414–5419
Tai M, Stephanopoulos G (2013) Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab Eng 15:1–9. doi:10.1016/j.ymben.2012.08.007
Tang W, Zhang S, Tan H, Zhao ZK (2010) Molecular cloning and characterization of a malic enzyme gene from the oleaginous yeast Lipomyces starkeyi. Mol Biotechnol 45:121–128
Thorpe RF, Ratledge C (1972) Fatty acid distribution in triglycerides of yeasts grown on glucose or n-Alkanes. Microbiology 72:151–163
Voegele RT, Mitsch MJ, Finan TM (1999) Characterization of two members of a novel malic enzyme class. Biochim Biophys Acta 1432:275–285
Wang JH, Hung W, Tsai SH (2011) High efficiency transformation by electroporation of Yarrowia lipolytica. J Microbiol 49:469–472
Wynn JP, Kendrick A, Ratledge C (1997) Sesamol as an inhibitor of growth and lipid metabolism in M. circinelloides via its action on malic enzyme. Lipids 32:605–610
Wynn JP, Hamid AA, Li Y, Ratledge C (2001) Biochemical events leading to the diversion of carbon into storage lipids in the oleaginous fungi M. circinelloides and M. alpina. Microbiology 147:2857–2864
Xue ZX, Sharpe PL, Hong SP, Yadav1 NS et al (2013) Sustainable source of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nature Biotech (in press). doi:10.1038/nbt.2622
Yu Q, Liu J, Wang Z, Nai J, Lü M, Zhou X, Cheng Y (2013) Characterization of the NADP-malic enzymes in the woody plant Populus trichocarpa. Mol Biol Rep 40:1385–1396
Zhang Y, Adams IP, Ratledge C (2007) Malic enzyme: the controlling activity for lipid production? Overexpression of malic enzyme in M. circinelloides leads to a 2.5-fold increase in lipid accumulation. Microbiology 153:2013–2025
Zhang B, Rong C, Chen H, Song Y, Zhang H, Chen W (2012a) De novo synthesis of trans-10, cis-12 conjugated linoleic acid in oleaginous yeast Yarrowia lipolytica. Microb Cell Fact 11:51. doi:10.1186/1475-2859-11-51
Zhang H, Damude HG, Yadav NS (2012b) Three diacylglycerol acyltransferases contribute to oil biosynthesis and normal growth in Yarrowia lipolytica. Yeast 29:25–38
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31271812,81071685,21276108), the National High Technology Research and Development Program of China (863 Program 2012AA022105C), the National Basic Research Program of China 973 Program (2012CB720802), the National Science Fund for Distinguished Young Scholars (31125021), the 111 project B07029, Starting Grant from Institut Mérieux (Strategic Mérieux Research Grants), and Institut Mérieux Fellowship 2012.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, H., Zhang, L., Chen, H. et al. Regulatory properties of malic enzyme in the oleaginous yeast, Yarrowia lipolytica, and its non-involvement in lipid accumulation. Biotechnol Lett 35, 2091–2098 (2013). https://doi.org/10.1007/s10529-013-1302-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-013-1302-7