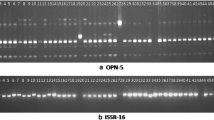
Camellia nitidissima, a rare plant but a useful genetic resource for commercial cultivation of ornamental camellias, is distributed in a narrow region of South China and North Vietnam. In this study, RAPD and AFLP markers were used to assess the genetic diversity and population structure of six natural populations of C. nitidissima from Guangxi in South China. Twenty RAPD primers amplified 183 bands, of which 143 bands were polymorphic, and 8 AFLP primer pairs produced 502 bands, of which 364 were polymorphic. Independent as well as combined analyses of the cluster analyses of the RAPD and AFLP fragments showed that the six populations could be classified into two major genetic groups corresponding to the Nanning and Fangcheng areas. The Mantel test revealed significant correlation between the genetic and geographic distances of C. nitidissima populations (r = 0.953, p = 0.036). AMOVA analysis allowed the partitioning of the genetic variation between groups (36.09%), among populations within groups (25.78%), and within populations (38.14%). An understanding of both the genetic diversity and the population structure of C. nitidissima in China can also provide insight into the conservation and management of this endangered species.


Similar content being viewed by others
REFERENCES
Cao, G. X., Zhong, Z. C., Xie, D. T., Liu, Y., and Long, Y. (2003). RAPD analysis of Camellia rosthorniana populations in different communities in Jinyun Mountain. Acta Ecol. Sin. 23:1583–1589.
Chang, H. T., and Ren, S. X. (1998). Flora Reipublicae Popularis Sinicae, vol. 49, 3rd edn., Science Press, Beijing, China, pp. 101–112.
Chung, M. G., and Chung M. Y. (2001). Levels and partitioning of genetic diversity of Camellia japonica (Theaceae) in Korea and Japon. Silvae Genet. 49:119-124.
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19:11–15.
Excoffier, L., Smouse, P. E., and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction sites. Genetics 131:474–479.
Fu, L. G. (1992). China Plant Red Data Book, Science Press, Beijing, China, pp. 648–649.
Godt, M. J. W., and Hamrick, J. L. (1993). Genetic diversity and population structure in Tradescantia hirsuticaulis (Commelinaceae). Am. J. Bot. 80:959–966.
Hamrick, J. L., Godt, M. J. W., Murawski, D. A., and Loveless, M. D. (1991). Correlations between species traits and allozyme diversity: Implications for conservation biology. In Falk, D. A., and Holsinger, K. E. (eds.), Genetics and Conservation of Rare Plants, Oxford University Press, New York, pp. 75–86.
Holsinger, K. E., and Gottlieb, L. D. (1991). Conservation of rare and endangered plants: Principles and prospects. In Falk, D. A., and Holsinger, K. E. (eds.), Genetics and Conservation of Rare Plants, Oxford University Press, New York, pp. 195–208.
Huang, F. P. (2001). Study on the yellow camellias community in Fangcheng. Guangxi Forest. Sci. 30:35–38.
Lewontin, R. C. (1972). The apportionment of human diversity. Evol. Biol. 6:381–398.
Liang, S. Y. (1993). Yellow Camellias, Chinese Forestry Press, Beijing, China, pp. 1–21.
Lynch, M., and Milligan, B. G. (1994). Analysis of population genetic structure with RAPD markers. Mol. Ecol. 3:91–99.
Mantel, N. A. (1967). The detection of disease clustering and a generalized regression approach. Cancer Res. 27:209–220.
Mueller, U. G., and Wolfenbarer, L. L. (1999). AFLP genotyping and fingerprinting. Trends Ecol. Evol. 14:389–394.
Nan, P., Shi, S., Peng, S., Tian, C., and Zhong, Y. (2003). Genetic diversity in Primula obconica from Central and South-west China as revealed by ISSR markers. Ann. Bot. 91:329–333.
Nei, M. (1973). Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. U.S.A. 70:3321–3323.
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590.
Nishimoto, S. I., Hashimoto, F., Shimizu, K., and Sakata, Y. (2004). Petal coloration of interspecific hydrids between Camellia chysantha × C. japonica. Jpn. Soc. Hort. Sci. 73:189–191.
Nybom, H., and Bartish, I. V. (2000). Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspec. Plant Ecol. Evol. Syst. 3(2):93–114.
Parks, C. R. (2000). Breeding progress with yellow camellias. American Camellia Yearbook, pp. 9–10.
Paul, S., Wchira, F. N., Powell, W., and Waugh, R. (1997). Diversity and genetic differentiation among populations of Indian and Kenyan tea (Camellia sinensis (L.) O. Kuntze) revealed by AFLP markers. Theor. Appl. Genet. 94:255–263.
Rohlf, F. J. (2000). NTSYSpc: Numerical taxonomy and multivariate analysis system, version 2.1, Exeter Software, Setauket, NY.
Schneider, S., Roessli, D., and Excoffier, L. (2000). Arlequin version 2000: A software for population genetics data analysis. University of Geneva.
Su, Z. M., and Mo, X. L. (1988). Geographic distribution of Camellia section Chrysantha from China. Guihaia 8:75–81.
Tang, S., Shi, S., Chen, Y., Qu, L., and Zhang, H. (1998). Phylogenetic relationship of Camellia nitidissima Chi and its allied species based on random amplified polymorphic DNA. Acta Sci. Nat. Univ. Sunyatseni 37(4):28–32.
Tang, T., Zhong, Y., Jian, S., and Shi, S. (2003). Genetic diversity of Hibiscus tiliaceus (Malvaceae) in China assessed using AFLP markers. Ann. Bot. 92:409–414.
Torres, E., Iriondo, J. M., and Perez, C. (2003). Genetic structure of an endangered plant, Antirrhinum microphyllum (Scrophulariaceae): Allozyme and RAPD analysis. Am. J. Bot. 90:85–92.
Vos, P., Hogers, R., and Bleeder, M. (1995). AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407–4414.
Wendel, J. F., and Parks, C. R. (1985). Genetic diversity and population structure in Camellia japonica L. (Theaceae). Am. J. Bot. 72:52–65.
Williams, J. G. K., Kubelik, A. R., Livak, K. J., Rafalski, J. A., and Tingey, S. V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531–6536.
Wright, S. (1931). Evolution in Mendelian populations. Genetics 16:97–159.
Wright, S. (1951). The genetical structure of populations. Ann. Engen. 15:323–354.
Yeeh, Y., Kang, S. S., and Chung, M. G. (1996). Evaluations of the natural monument populations of Camellia japonica (Theaceae) in Korea based on allozyme studies. Bot. Bull. Acad. Sin. 37:141–146.
Yeh, F. C., Yang, R., Boyle, T. J., Ye, Z., and Xiyan, J. M. (2000). PopGene32, Microsoft Windows-based freeware for population. Genetic analysis, version 1.32, Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton.
Zietkiewicz, E., Rafalski, A., and Labuda, D. (1994). Genome fingerprinting by simple sequence repeats (SSR)-anchored PCR amplification. Genomics 20:176–183.
ACKNOWLEDGMENTS
We would like to thank Jimei Liu and Wenjuan Zhang for technical assistance. This work was supported by grants from the Chinese National Natural Science Foundation (30260013), the program for NCET in university and Chinese National Key Project for Basic Research (973) (2002CB512801).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tang, S., Bin, X., Wang, L. et al. Genetic Diversity and Population Structure of Yellow Camellia (Camellia nitidissima) in China as Revealed by RAPD and AFLP Markers. Biochem Genet 44, 444–456 (2006). https://doi.org/10.1007/s10528-006-9053-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-006-9053-y