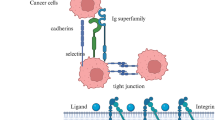
Abstract
During cancer cell invasion, integrin undergoes constant endo/exocytic trafficking. It has been found that the recycling ability of integrin β1 through Rab11-controlled long loop pathways is directly associated with cancer invasion. Previous studies showed that gain-of-function mutant p53 regulates the Rab-coupling protein [RCP]-mediated integrin β1 recycling by inactivating tumor suppressor TAp63. So, we were interested to investigate the involvement of miR-205 in this process. In the current study first, we evaluated that the lower expression of miR-205 in MDA-MB-231 cell line is associated with high motility and invasiveness. Further investigation corroborated that miR-205 directly targets RCP resulting in attenuated RCP-mediated integrin β1 recycling. Overexpression of TAp63 validates our in vitro findings. To appraise the anti-metastatic role of miR-205, we developed two in vivo experimental models- xenograft-chick embryo and xenograft-immunosuppressed BALB/c mice. Our in vivo results support the negative effect of miR-205 on metastasis. Therefore, these findings advocate the tumor suppressor activity of miR-205 in breast cancer cells and suggest that in the future development of miR-205-targeting RNAi therapeutics could be a smart alternative approach to prevent the metastatic fate of the disease.








Similar content being viewed by others
Data Availability
Privacy/ethical restrictions: The raw data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
References
Hapach LA, Mosier JA, Wang W, Reinhart-King CA (2019) Engineered models to parse apart the metastatic cascade. Npj Precision Oncology 3(1):20. https://doi.org/10.1038/s41698-019-0092-3
Lauffenburger DA, Horwitz AF (1996) Cell Migration: a physically integrated molecular process. Cell 84(3):359–369. https://doi.org/10.1016/S0092-8674(00)81280-5
Sheetz MP (1994) Cell migration by graded attachment to substrates and contraction. Semin Cell Biol 5(3):149–155. https://doi.org/10.1006/scel.1994.1019
Lee J, Ishihara A, Theriot JA, Jacobson K (1993) Principles of locomotion for simple-shaped cells. Nature 362(6416):167–171. https://doi.org/10.1038/362167a0
Webb DJ, Parsons JT, Horwitz AF (2002) Adhesion assembly, disassembly and turnover in migrating cells – over and over and over again. Nat Cell Biol 4(4):E97–E100. https://doi.org/10.1038/ncb0402-e97
Hynes RO, Integrins (2002) Bidirectional, Allosteric Signaling Machines. Cell 110(6):673–687. https://doi.org/10.1016/S0092-8674(02)00971-6
Caswell PT, Norman JC (2006) Integrin trafficking and the control of Cell Migration. Traffic 7(1):14–21. https://doi.org/10.1111/j.1600-0854.2005.00362.x
Jones MC, Caswell PT, Norman JC (2006) Endocytic recycling pathways: emerging regulators of cell migration. Curr Opin Cell Biol 18(5):549–557. https://doi.org/10.1016/j.ceb.2006.08.003
Bridgewater RE, Norman JC, Caswell PT (2012) Integrin trafficking at a glance. J Cell Sci 125(16):3695–3701. https://doi.org/10.1242/jcs.095810
De Franceschi N, Hamidi H, Alanko J, Sahgal P, Ivaska J (2015) Integrin traffic – the update. J Cell Sci 128(5):839–852. https://doi.org/10.1242/jcs.161653
Parachoniak CA, Park M (2012) Dynamics of receptor trafficking in tumorigenicity. Trends Cell Biol 22(5):231–240. https://doi.org/10.1016/j.tcb.2012.02.002
White DP, Caswell PT, Norman JC (2007) αvβ3 and α5β1 integrin recycling pathways dictate downstream rho kinase signaling to regulate persistent cell migration. J Cell Biol 177(3):515–525. https://doi.org/10.1083/jcb.200609004
Fang Z, Takizawa N, Wilson KA, Smith TC, Delprato A, Davidson MW et al (2010) The membrane-Associated protein, Supervillin, accelerates F-Actin-dependent Rapid Integrin Recycling and Cell Motility. Traffic 11(6):782–799. https://doi.org/10.1111/j.1600-0854.2010.01062.x
Powelka AM, Sun J, Li J, Gao M, Shaw LM, Sonnenberg A et al (2004) Stimulation-dependent recycling of integrin β1 regulated by ARF6 and Rab11. Traffic 5(1):20–36. https://doi.org/10.1111/j.1600-0854.2004.00150.x
Arjonen A, Alanko J, Veltel S, Ivaska J (2012) Distinct recycling of active and inactive β1 integrins. Traffic 13(4):610–625. https://doi.org/10.1111/j.1600-0854.2012.01327.x
Goldenring JR, Ray GS, Lee JR (1999) Rab11 in dysplasia of Barrett’s epithelia. Yale J Biol Med. ;72(2–3):113 – 20. Epub 2000/04/26. PubMed PMID: 10780572; PubMed Central PMCID: PMCPMC2579020
Caswell PT, Chan M, Lindsay AJ, McCaffrey MW, Boettiger D, Norman JC (2008) Rab-coupling protein coordinates recycling of α5β1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J Cell Biol 183(1):143–155. https://doi.org/10.1083/jcb.200804140
Muller PAJ, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S et al (2009) Mutant p53 drives Invasion by promoting integrin recycling. Cell 139(7):1327–1341. https://doi.org/10.1016/j.cell.2009.11.026
Muller PAJ, Trinidad AG, Timpson P, Morton JP, Zanivan S, van den Berghe PVE et al (2013) Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene 32(10):1252–1265. https://doi.org/10.1038/onc.2012.148
Zhang J, Liu X, Datta A, Govindarajan K, Tam WL, Han J et al (2009) RCP is a human Breast cancer–promoting gene with ras-activating function. J Clin Investig 119(8):2171–2183. https://doi.org/10.1172/JCI37622
Balsara BR, Sonoda G, du Manoir S, Siegfried JM, Gabrielson E, Testa JR (1997) Comparative genomic hybridization analysis detects frequent, often High-Level, overrepresentation of DNA sequences at 3q, 5p, 7p, and 8q in human non-small cell lung carcinomas. Cancer Res 57(11):2116–2120
Dai Y, Liu Y, Huang D, Yu C, Cai G, Pi L et al (2012) Increased expression of Rab coupling protein in squamous cell carcinoma of the head and neck and its clinical significance. Oncol Lett 3(6):1231–1236 Epub 2012/07/12. https://doi.org/10.3892/ol.2012.652
Mills GB, Jurisica I, Yarden Y, Norman JC (2009) Genomic amplicons target vesicle recycling in Breast cancer. J Clin Investig 119(8):2123–2127. https://doi.org/10.1172/JCI40256
Muller Patricia AJ, Vousden Karen H (2014) Mutant p53 in Cancer: New functions and Therapeutic opportunities. Cancer Cell 25(3):304–317. https://doi.org/10.1016/j.ccr.2014.01.021
Varna M, Bousquet G, Plassa L-F, Bertheau P, Janin A (2011) TP53 status and response to treatment in breast cancers. J Biomed Biotechnol 2011:284584. https://doi.org/10.1155/2011/284584
Neilsen PM, Noll JE, Mattiske S, Bracken CP, Gregory PA, Schulz RB et al (2013) Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene 32(24):2992–3000. https://doi.org/10.1038/onc.2012.305
Muller PAJ, Vousden KH, Norman JC (2011) p53 and its mutants in Tumor cell migration and invasion. J Cell Biol 192(2):209–218. https://doi.org/10.1083/jcb.201009059
Muller PAJ, Trinidad AG, Caswell PT, Norman JC, Vousden KH (2014) Mutant p53 regulates dicer through p63-dependent and -independent mechanisms to promote an invasive phenotype. J Biol Chem 289(1):122–132. https://doi.org/10.1074/jbc.M113.502138
Li XL, Jones MF, Subramanian M, Lal A (2014) Mutant p53 exerts oncogenic effects through microRNAs and their target gene networks. FEBS Lett 588(16):2610–2615. https://doi.org/10.1016/j.febslet.2014.03.054
Muller PAJ, Vousden KH (2013) p53 mutations in cancer. Nat Cell Biol 15(1):2–8. https://doi.org/10.1038/ncb2641
Roger L, Jullien L, Gire V, Roux P (2010) Gain of oncogenic function of p53 mutants regulates E-cadherin expression uncoupled from cell invasion in colon Cancer cells. J Cell Sci 123(8):1295–1305. https://doi.org/10.1242/jcs.061002
Li Y, Prives C (2007) Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene 26(15):2220–2225. https://doi.org/10.1038/sj.onc.1210311
Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C (2001) A subset of Tumor-Derived Mutant forms of p53 down-regulate p63 and p73 through a Direct Interaction with the p53 core domain. Mol Cell Biol 21(5):1874–1887. https://doi.org/10.1128/mcb.21.5.1874-1887.2001
Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/Smad Complex Opposes p63 to Empower TGFβ-Induced Metastasis. Cell. 2009;137(1):87–98. https://doi.org/10.1016/j.cell.2009.01.039
Girardini Javier E, Napoli M, Piazza S, Rustighi A, Marotta C, Radaelli E et al (2011) A Pin1/Mutant p53 Axis Promotes Aggressiveness in Breast Cancer. Cancer Cell 20(1):79–91. https://doi.org/10.1016/j.ccr.2011.06.004
Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin Y-L et al (2010) TAp63 suppresses Metastasis through coordinate regulation of Dicer and miRNAs. Nature 467(7318):986–990. https://doi.org/10.1038/nature09459
Tran MN, Choi W, Wszolek MF, Navai N, Lee I-LC, Nitti G et al (2013) The p63 protein isoform ∆Np63α inhibits epithelial-mesenchymal transition in human Bladder Cancer cells: ROLE OF MIR-205. J Biol Chem 288(5):3275–3288. https://doi.org/10.1074/jbc.M112.408104
Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vousden KH et al (2012) Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proceedings of the National Academy of Sciences. ;109(38):15312-7. https://doi.org/10.1073/pnas.1110977109
Xiao Y, Humphries B, Yang C, Wang Z (2019) MiR-205 dysregulations in Breast Cancer: the Complexity and opportunities. Non-Coding RNA 5(4):53 PubMed PMID. https://doi.org/10.3390/ncrna5040053
Qin A-Y, Zhang X-W, Liu L, Yu J-P, Li H, Emily Wang S-Z et al (2013) MiR-205 in cancer: an angel or a devil? Eur J Cell Biol 92(2):54–60. https://doi.org/10.1016/j.ejcb.2012.11.002
Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T et al (2009) microRNA-205 regulates HER3 in human Breast Cancer. Cancer Res 69(6):2195–2200. https://doi.org/10.1158/0008-5472.can-08-2920
Wu H, Zhu S, Mo Y-Y (2009) Suppression of cell growth and invasion by miR-205 in Breast cancer. Cell Res 19(4):439–448. https://doi.org/10.1038/cr.2009.18
Piovan C, Palmieri D, Di Leva G, Braccioli L, Casalini P, Nuovo G et al (2012) Oncosuppressive role of p53-induced miR-205 in triple negative Breast cancer. Mol Oncol 6(4):458–472. https://doi.org/10.1016/j.molonc.2012.03.003
Elgamal OA, Park J-K, Gusev Y, Azevedo-Pouly ACP, Jiang J, Roopra A et al (2013) Tumor suppressive function of mir-205 in Breast Cancer is linked to HMGB3 regulation. PLoS ONE 8(10):e76402. https://doi.org/10.1371/journal.pone.0076402
Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G et al (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10(5):593–601. https://doi.org/10.1038/ncb1722
Lee JY, Park MK, Park JH, Lee HJ, Shin DH, Kang Y et al (2014) Loss of the polycomb protein Mel-18 enhances the epithelial–mesenchymal transition by ZEB1 and ZEB2 expression through the downregulation of miR-205 in Breast cancer. Oncogene 33(10):1325–1335. https://doi.org/10.1038/onc.2013.53
Kim ES, Choi JY, Hwang SJ, Bae IH (2019) Hypermethylation of mir-205-5p by IR governs aggressiveness and Metastasis via regulating Bcl-w and Src. Mol Therapy - Nucleic Acids 14:450–464. https://doi.org/10.1016/j.omtn.2018.12.013
Wang T, Zhao N, Long S, Ge L, Wang A, Sun H et al (2016) Downregulation of miR-205 in migrating epithelial tongue facilitates skin wound re-epithelialization by derepressing ITGA5. Biochimica et Biophysica Acta (BBA) - molecular basis of Disease. 1862(8):1443–1452. https://doi.org/10.1016/j.bbadis.2016.05.004
Xiao Y, Li Y, Tao H, Humphries B, Li A, Jiang Y et al (2018) Integrin α5 down-regulation by miR-205 suppresses triple negative Breast cancer stemness and Metastasis by inhibiting the Src/Vav2/Rac1 pathway. Cancer Lett 433:199–209. https://doi.org/10.1016/j.canlet.2018.06.037
Roberts M, Barry S, Woods A, van der Sluijs P, Norman J (2001) PDGF-regulated rab4-dependent recycling of avb3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr Biol 11(18):1392–1402. https://doi.org/10.1016/S0960-9822(01)00442-0
Bhattacharya S, Ghosh A, Maiti S, Ahir M, Debnath GH, Gupta P et al (2020) Delivery of thymoquinone through hyaluronic acid-decorated mixed Pluronic® nanoparticles to attenuate angiogenesis and Metastasis of triple-negative Breast cancer. J Controlled Release 322:357–374. https://doi.org/10.1016/j.jconrel.2020.03.033
Kim S, Shephard N, Chib S (1998) Stochastic volatility: Likelihood Inference and comparison with ARCH models. Rev Econ Stud 65(3):361–393
Upadhyay P, Sarker S, Ghosh A, Gupta P, Das S, Ahir M et al (2019) Transferrin-decorated thymoquinone-loaded PEG-PLGA nanoparticles exhibit anticarcinogenic effect in non-small cell lung carcinoma via the modulation of miR-34a and miR-16. Biomaterials Sci 7(10):4325–4344. https://doi.org/10.1039/C9BM00912D
Akao Y, Nakagawa Y, Naoe T (2006) MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep 16(4):845–850 Epub 2006/09/14. PubMed PMID: 16969504
Peng Y, Croce CM (2016) The role of MicroRNAs in human cancer. Signal Transduct Target Therapy 1(1):15004. https://doi.org/10.1038/sigtrans.2015.4
Schwerk J, Savan R (2015) Translating the untranslated region. J Immunol 195(7):2963–2971. https://doi.org/10.4049/jimmunol.1500756PubMed PMID: 26386038
Luo D, Wilson JM, Harvel N, Liu J, Pei L, Huang S et al (2013) A systematic evaluation of miRNA:mRNA interactions involved in the migration and invasion of Breast cancer cells. J Translational Med 11(1):57. https://doi.org/10.1186/1479-5876-11-57
Plantamura I, Cataldo A, Cosentino G, Iorio MV (2020) miR-205 in Breast Cancer: state of the art. Int J Mol Sci 22(1):27. https://doi.org/10.3390/ijms22010027PubMed PMID: 33375067
Haleh V, Ali S, Robert Anthony S, Alfred King-Yin L (2014) The important roles of miR-205 in normal physiology, cancers and as a potential therapeutic target. Curr Cancer Drug Targets 14(7):621–637. https://doi.org/10.2174/156800961407140926105634
Greene SB, Herschkowitz JI, Rosen JM (2010) The ups and downs of miR-205: identifying the roles of miR-205 in mammary gland development and Breast cancer. RNA Biol 7(3):300–304. https://doi.org/10.4161/rna.7.3.11837
Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G et al (2007) Altered MicroRNA expression confined to specific epithelial cell subpopulations in Breast Cancer. Cancer Res 67(24):11612–11620. https://doi.org/10.1158/0008-5472.can-07-5019
Berber U, Yilmaz I, Narli G, Haholu A, Kucukodaci Z, Demirel D (2014) miR-205 and miR-200c: Predictive Micro RNAs for Lymph Node Metastasis in Triple negative Breast Cancer. J Breast Cancer 17(2):143–148
Du Y-e, Tu G, Yang G, Li G, Yang D, Lang L et al (2017) MiR-205/YAP1 in activated fibroblasts of breast Tumor promotes VEGF-independent angiogenesis through STAT3 signaling. Theranostics 7(16):3972–3988. https://doi.org/10.7150/thno.18990
Caswell PT, Spence HJ, Parsons M, White DP, Clark K, Cheng KW et al (2007) Rab25 Associates with 51 Integrin to Promote Invasive Migration in 3D Microenvironments. Dev Cell 13(4):496–510. https://doi.org/10.1016/j.devcel.2007.08.012
Hales CM, Griner R, Hobdy-Henderson KC, Dorn MC, Hardy D, Kumar R et al (2001) Identification and characterization of a family of Rab11-interacting proteins. J Biol Chem 276(42):39067–39075. https://doi.org/10.1074/jbc.M104831200
Lindsay AJ, McCaffrey MW (2002) Rab11-FIP2 functions in Transferrin Recycling and Associates with endosomal membranes via its COOH-terminal domain. J Biol Chem 277(30):27193–27199. https://doi.org/10.1074/jbc.M200757200
Lindsay AJ, Hendrick AG, Cantalupo G, Senic-Matuglia F, Goud B, Bucci C et al (2002) Rab Coupling Protein (RCP), a Novel Rab4 and Rab11 effector protein. J Biol Chem 277(14):12190–12199. https://doi.org/10.1074/jbc.M108665200
Eva R, Dassie E, Caswell PT, Dick G, ffrench-Constant C, Norman JC et al (2010) Rab11 and its effector Rab coupling protein contribute to the trafficking of beta 1 integrins during axon growth in adult dorsal root ganglion neurons and PC12 cells. J Neurosci 30(35):11654–11669. https://doi.org/10.1523/JNEUROSCI.2425-10.2010PubMed PMID: 20810886
Caswell PT, Chan M, Lindsay AJ, McCaffrey MW, Boettiger D, Norman JC (2008) Rab-coupling protein coordinates recycling of alpha5beta1 integrin and EGFR1 to promote cell migration in 3D microenvironments. J Cell Biol 183(1):143–155. https://doi.org/10.1083/jcb.200804140PubMed PMID: 18838556
Gundry C, Marco S, Rainero E, Miller B, Dornier E, Mitchell L et al (2017) Phosphorylation of rab-coupling protein by LMTK3 controls Rab14-dependent EphA2 trafficking to promote cell:cell repulsion. Nat Commun 8:14646. https://doi.org/10.1038/ncomms14646PubMed PMID: 28294115
Peden AA, Schonteich E, Chun J, Junutula JR, Scheller RH, Prekeris R (2004) The RCP-Rab11 complex regulates endocytic protein sorting. Mol Biol Cell 15(8):3530–3541. https://doi.org/10.1091/mbc.e03-12-0918PubMed PMID: 15181150
Zhang J, Liu X, Datta A, Govindarajan K, Tam WL, Han J et al (2009) RCP is a human breast cancer-promoting gene with ras-activating function. J Clin Invest 119(8):2171–2183 Epub 07/20. doi: 10.1172/JCI37622. PubMed PMID: 19620787
Tang BL (2010) Is Rab25 a Tumor promoter or suppressor–context dependency on RCP status? Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine 31(4):359–361. https://doi.org/10.1007/s13277-010-0030-z. Epub 2010/04/09. PubMed PMID: 20376596
Hwang MH, Cho KH, Jeong KJ, Park YY, Kim JM, Yu SL et al (2017) RCP induces slug expression and cancer cell invasion by stabilizing β1 integrin. Oncogene 36(8):1102–1111 Epub 2016/08/16. https://doi.org/10.1038/onc.2016.277
Xiao Y, Li Y, Tao H, Humphries B, Li A, Jiang Y et al (2018) Integrin α5 down-regulation by miR-205 suppresses triple negative Breast cancer stemness and Metastasis by inhibiting the Src/Vav2/Rac1 pathway. Cancer Lett 433:199–209 Epub 2018/07/03. https://doi.org/10.1016/j.canlet.2018.06.037
Croissant JD, Carpenter S, Bader D (2000) Identification and genomic cloning of CMHC1. A unique myosin heavy chain expressed exclusively in the developing chicken heart. J Biol Chem 275(3):1944–1951 Epub 2000/01/15. https://doi.org/10.1074/jbc.275.3.1944
Acknowledgements
We would like to acknowledge Department of Biotechnology [Sanction no. 6242-P23/RGCB/PMD/DBT/ARAD-2015 dated 29th July 2015], Council of Scientific and Industrial Research, India [CSIR Sanction no. 09/028[0966]/2016-EMR-I]; DST-NANOMISSION, India [SR/NM/NS-1185/2015[g] ; DBT-Ayush India, [sanction no: Z.28015/29/2016-HPC [EMR]-AYUSH-A], DST INSPIRE Faculty Scheme [sanction no: IFA12-LSBM-40], Govt. of India and DST-INSPIRE, Department of Science and Technology, for funding support. Authors would like to thank Mr. Abir Chakraborty and Dr. Sneha Mitra for their technical support.
Author information
Authors and Affiliations
Contributions
Saurav Bhattacharya: Conceptualization, methodology, data leading curation, investigation, writing - original draft. Sushmita Sarker: Methodology, formal Analysis, data curation. Shaswati Das: Formal analysis, data curation. Manisha Ahir: Formal analysis, data curation.: Sreya Chattopadhyay: Formal analysis. Swatilekha Ghosh: Formal analysis, writing review & editing. Arghya Adhikary: Conceptualization, supervision, project administration, funding acquisition, writing - review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors report no conflicts of interest in this work.
Included in article
The representative data that support the findings of this study are available in the methods and/or supplementary material of this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bhattacharya, S., Sarker, S., Das, S. et al. microRNA-205 represses breast cancer metastasis by perturbing the rab coupling protein [RCP]-mediated integrin β1 recycling on the membrane. Apoptosis 29, 191–209 (2024). https://doi.org/10.1007/s10495-023-01912-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-023-01912-7