Abstract
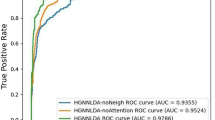
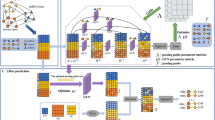
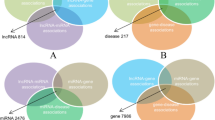
The advancements in the field of high throughput analysis show abnormal expression of long non-coding RNAs (lncRNAs) in many complex diseases. Accurately identifying the disease association of lncRNA is essential in understanding their role in disease mechanism and subsequent therapy. The contemporary methods for predicting lncRNA-disease association use heterogeneous information learned from different biological sources such as lncRNAs, miRNAs, and diseases. However, learning topological features from diverse network structured data is one of the limiting factors of these methods. To address this challenge, we propose a method for lncRNA-disease association prediction based on Deep Belief Network (DBN), referred to as DBNLDA. In this method, three interaction networks such as lncRNA-miRNA similarity (LMS), disease-miRNA similarity (DMS), and lncRNA-disease association (LDA) network are constructed. A new framework based on the node embedding, DBN, and a neural network regression model is used to learn network and local representation of lncRNA-disease pairs. From the node embedding matrices of LMS, DMS, and LDA networks, lncRNA-disease features are learned by DBN layers. These DBN features are used to predict the association score by an ANN regression model. Compared to several state-of-the-art methods, DBNLDA obtained better AUC (0.96) and AUPR (0.967) under five-fold cross-validation. Case studies on breast, lung, and stomach cancer also affirmed the ability of DBNLDA in predicting potential lncRNAs associated with various diseases.




Similar content being viewed by others
References
Bahari F, Emadi-Baygi M, Nikpour P et al (2015) miR-17-92 host gene, uderexpressed in gastric cancer and its expression was negatively correlated with the metastasis. Ind J Cancer 52(1):22
Bhartiya D, Kapoor S, Jalali S, Sati S, Kaushik K, Sachidanandan C, Sivasubbu S, Scaria V (2012) Conceptual approaches for lncRNA drug discovery and future strategies. Expert Opin Drug Discov 7(6):503–513
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, Zhang Q, Yan G, Cui Q (2012) LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res 41(D1):D983–D986
Chen X (2015) KATZLDA: KATZ Measure for the lncRNA-disease association prediction. Sci Rep 5:16840
Chen X, Yan CC, Luo C, Ji W, Zhang Y, Dai Q (2015) Constructing lncRNA functional similarity network based on lncRNA-disease associations and disease semantic similarity. Sci Rep 5:11338
Chen X, Yan CC, Luo C, Ji W, Zhang Y, Dai Q (2015) Constructing lncRNA functional similarity network based on lncRNA-disease associations and disease semantic similarity. Sci Rep 5:11338
Chen X, Yan CC, Zhang X, Li Z, Deng L, Zhang Y, Dai Q (2015) RBMMMDA: Predicting multiple types of disease-microRNA associations. Sci Rep 5:13877
Chen X, You Z-H, Yan G-Y, Gong D-W (2016) IRWRLDA: Improved random walk with restart for lncRNA-disease association prediction. Oncotarget 7(36):57919
Davis J, Goadrich M (2006) The relationship between precision-recall and roc curves. In: Proceedings of the 23rd international conference on Machine learning, pp 233–240
Ding L, Wang M, Sun D, Li A (2018) TPGLDA Novel Prediction of associations between lncRNAs and diseases via lncRNA-disease-gene tripartite graph. Sci Rep 8(1):1–11
Du Y, Hao X, Liu X (2018) Low expression of long noncoding rna cdkn2b-as1 in patients with idiopathic pulmonary fibrosis predicts lung cancer by regulating the p53-signaling pathway. Oncol Lett 15(4):4912–4918
Fan Y, Siklenka K, Arora SK, Ribeiro P, Kimmins S, Xia J (2016) miRNet-dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res 44 (W1):W135–W141
Fu G, Wang J, Domeniconi C, Yu G (2018) Matrix factorization-based data fusion for the prediction of lncRNA–disease associations. Bioinformatics 34(9):1529–1537
Goff LA, Rinn JL (2015) Linking RNA biology to lncRNAs. Genome Res 25(10):1456–1465
Grover A, Leskovec J (2016) node2vec Scalable feature learning for networks. In: Proceedings of the 22nd ACM SIGKDD international conference on Knowledge discovery and data mining, pp 855–864
Guo S, Zhang L, Zhang Y, Wu Z, He D, Li X, Wang Z (2019) Long non-coding RNA TUG1 enhances chemosensitivity in non-small cell lung cancer by impairing microRNA-221-dependent PTEN inhibition. Aging (Albany NY) 11(18):7553
Hinton GE (2009) Deep belief networks. Scholarpedia 4(5):5947
Huarte M (2015) The emerging role of lncRNAs in cancer. Nat Med 21(11):1253
Kipf TN, Welling M (2016) Semi-supervised classification with graph convolutional networks. arXiv:1609.02907
Lan W, Li M, Zhao K, Liu J, Wu F-X, Yi P, Wang J (2017) LDAP: A web server for lncRNA-disease association prediction. Bioinformatics 33(3):458–460
Lee JT (2012) Epigenetic regulation by long noncoding RNAs. Science 338(6113):1435–1439
Li JW, Gao C, Wang Y, Ma W, Tu J, Wang JP, Chen Z, Kong W, Cui Q (2014) A bioinformatics method for predicting long noncoding rnas associated with vascular disease. Sci China Life Sci 57(8):852–857
Li J-H, Liu S, Zhou H, Qu L-H, Yang J-H (2014) starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res D1:D92–D97
Liang M, Li Z, Chen T, Zeng J (2014) Integrative data analysis of multi-platform cancer data with a multimodal deep learning approach. IEEE/ACM Trans Comput Biol Bioinform 12(4):928–937
L Liu X Y, Zhou J Q, Zhang G, Wang J, He Y, Chen C, Huang L (2018) Li, and SQ Li. LncRNA HULC promotes non-small cell lung cancer cell proliferation and inhibits the apoptosis by up-regulating sphingosine kinase 1 (SPHK1) and its downstream PI3k/akt pathway. Eur Rev Med Pharmacol Sci 22:8722–8730
Liu M-X, Chen X, Chen G, Cui Q-H, Yan G-Y (2014) A computational framework to infer human disease-associated long noncoding rnas. PloS One 9(1):e84408
Lu C, Yang M, Luo F, Wu F-X, Li M, Yi P, Li Y, Wang J (2018) Prediction of lncRNA–disease associations based on inductive matrix completion. Bioinformatics 34(19):3357–3364
Luo P, Li Y, Tian L-P, Wu F-X (2019) Enhancing the prediction of disease–gene associations with multimodal deep learning. Bioinformatics 35(19):3735–3742
Luo Q, Chen Y (2016) Long noncoding RNAs and Alzheimer’s disease. Clin Intervent Aging 11:867
Ning S, Zhang J, Wang P, Zhi H, Wang J, Liu Y, Gao Y, Guo M, Yue M, Wang L et al (2016) Lnc2cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res 44(D1):D980–D985
Paraskevopoulou MD, Hatzigeorgiou AG (2016) Analyzing miRNA–lncRNA interactions. In: long non-coding RNAs. Springer, pp 271–286
Piao H-Y, Guo S, Wang Y, Zhang J (2019) Long noncoding RNA NALT1-induced gastric cancer invasion and metastasis via NOTCH signaling pathway. World J Gastroenterol 25(44):6508
Ping P, Wang L, Kuang L, Ye S, Iqbal MFB, Pei T (2018) A novel method for lncRNA-disease association prediction based on an lncRNA-disease association network. IEEE/ACM Trans Comput Biol Bioinform 16(2):688–693
Ren H, Yang X, Yang Y, Zhang X, Zhao R, Wei R, Zhang X, Yi Z (2018) Upregulation of lncRNA BCYRN1 promotes tumor progression and enhances epCAM expression in gastric carcinoma. Oncotarget 9(4):4851
Song J, Wang Q, Zong L (2020) LncRNA MIR155HG contributes to smoke-related chronic obstructive pulmonary disease by targeting miR-128-5p/BRD4 axis. Biosci Rep 40(3)
Sun J, Shi H, Wang Z, Zhang C, Liu L, Wang L, He W, Hao D, Liu S, Zhou M (2014) Inferring novel lncRNA–disease associations based on a random walk model of a lncRNA functional similarity network. Mol BioSyst 10(8):2074–2081
Wang G, Zheng X, Zheng Y, Cao R, Zhang M, Sun Y, Wu J (2019) Construction and analysis of the lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveals functional genes in heart failure. Mol Med Rep 19(2):994–1003
Wang H, Shen Q, Zhang X, Yang C, Su C, Sun Y, Wang L, Fan X, Xu S (2017) The long non-coding RNA XIST controls non-small cell lung cancer proliferation and invasion by modulating miR-186-5p. Cell Physiol Biochem 41(6):2221–2229
Wang P, Guo Q, Gao Y, Zhi H, Zhang Y, Liu Y, Zhang J, Yue M, Guo M, Ning S et al (2017) Improved method for prioritization of disease associated lncRNAs based on ceRNA theory and functional genomics data. Oncotarget 8(3):4642
Wang S, Ke H, Zhang H, Ma Y, Ao L, Zou L, Yang Q, Zhu H, Nie J, Wu C et al (2018) LncRNA MIR100HG promotes cell proliferation in triple-negative breast cancer through triplex formation with p27 loci. Cell Death Disease 9(8):1–11
Wang Y, Zeng J (2013) Predicting drug-target interactions using restricted Boltzmann machines. Bioinformatics 29(13):i126–i134
Wu J, Chen H, Ye M, Wang B, Zhang Y, Sheng J, Meng T, Chen H (2019) Downregulation of long noncoding RNA HCP5 contributes to cisplatin resistance in human triple-negative breast cancer via regulation of PTEN expression. Biomed Pharmacother 115:108869
Wu X, Lan W, Chen Q, Dong Y, Liu J, Peng W (2020) Inferring LncRNA-disease Associations Based: On Graph Autoencoder Matrix Completion. Computational Biology and Chemistry, pp 107282
Xuan P, Cao Y, Zhang T, Kong R, Zhang Z (2019) Dual convolutional neural networks with attention mechanisms based method for predicting disease - related lncRNA genes. Front Gen 10:416
Xuan P, Pan S, Zhang T, Liu Y, Sun H (2019) Graph Convolutional Network and Convolutional Neural Network Based Method for Predicting lncRNA-disease Associations. Cells 8(9):1012
Yao D, Zhan X, Zhan X, Kwoh CK, Li P, Wang J (2020) A random forest based computational model for predicting novel lncRNA-disease associations. BMC Bioinform 21(1):1–18
Yu G, Fu G, Lu C, Ren Y, Wang J (2017) BRWLDA: Bi-random walks for predicting lncRNA-disease associations. Oncotarget 8(36):60429
Han J, Kamber M, Pei J (2011) Data mining concepts and techniques. The Morgan Kaufmann Series in Data Management Systems, pp 5.4
Acknowledgements
This research work is an outcome of the R&D work under the Visvesvaraya PhD Scheme of Ministry of Electronics and Information Technology, Government of India
Author information
Authors and Affiliations
Contributions
Both authors contributed equally in conception, design and implementation of the proposed idea and manuscript preparation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Supplementary Information
The online version contains supplementary material available at https://doi.org/10.1007/s10489-021-02675-x.
Availability of data and material
https://github.com/manumad/DBNLDA
Code availability
https://github.com/manumad/DBNLDA
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Madhavan, M., Gopakumar, G. DBNLDA: Deep Belief Network based representation learning for lncRNA-disease association prediction. Appl Intell 52, 5342–5352 (2022). https://doi.org/10.1007/s10489-021-02675-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10489-021-02675-x