Abstract
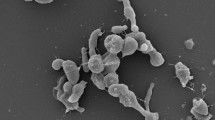
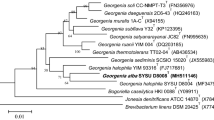
An orange-black, Gram-positive, aerobic and gamma-ray resistant actinobacterium was isolated from the ruins of a Roman aqueduct located in Northern Tunisia. The optimal growth for the strain was found to be at 25–35 °C and at pH 6.0–9.5. Chemotaxonomic and molecular characteristics of the isolate matched those described for members of the genus Geodermatophilus. The peptidoglycan was found to contain meso-diaminopimelic acid as diagnostic diaminoacid. The main polar lipids were identified as phosphatidylcholine, diphosphatidylglycerol, phosphatidylinositol, phosphatidylethanolamine, an unidentified glycolipid and an unidentified aminophospholipid; MK-9(H4) was found to be the dominant menaquinone and galactose was detected as the diagnostic sugar, with glucose, ribose and mannose also present. The major cellular fatty acids were identified as branched-chain saturated acids iso-C16:0, iso-C15:0 and iso-H-C16:0. The 16S rRNA gene showed 95.4–99.6 % sequence identity with the type strains of the genus Geodermatophilus. DNA–DNA relatedness values with closely related species were 39.9 ± 4.9, 33.9 ± 1.9, 27.0 ± 2.5 and 13.2 ± 1.35 % with Geodermatophilus amargosae, G. normandii, G. saharensis and G. tzadiensis respectively. Based on phenotypic results and 16S rRNA gene sequence analysis, strain BMG801T (=DSM 46834T = CECT 8822T) is proposed to represent the type strain of a novel species, Geodermatophilus aquaeductus sp. nov.



Similar content being viewed by others
References
Ahrens R, Moll G (1970) Ein neues knospendes Bakterium aus der Ostsee. Arch Mikrobiol 70:243–265
Battista JR, Earl AM, Park M-J (1999) Why is Deinococcus radiodurans so resistant to ionizing radiation? Trends Microbiol 7:362–365
Bertazzo M, Montero-Calasanz MC, Martinez-Garcia M et al (2014) Geodermatophilus brasiliensis sp. nov., isolated from Brazilian soil. Int J Syst Evol Microbiol 64:2841–2848
Cashion P, Holder-Franklin MA, McCully J, Franklin M (1977) A rapid method for the base ratio determination of bacterial DNA. Anal Biochem 81:461–466
Daly MJ (2009) A new perspective on radiation resistance based on Deinococcus radiodurans. Nat Rev Microbiol 7:237–245
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
Essoussi I, Ghodhbane-Gtari F, Amairi H et al (2010) Esterase as an enzymatic signature of Geodermatophilaceae adaptability to Sahara desert stones and monuments. J Appl Microbiol 108:1723–1732
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Fitch WM (1971) toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Gordon RE, Smith MM (1955) Proposed group of characters for the separation of Streptomyces and Nocardia. J Bacteriol 69:147–150
Gregersen T (1978) Rapid method for distinction of gram-negative from gram-positive bacteria. Eur J Appl Microbiol Biotechnol 5:123–127
Gtari M, Essoussi I, Maaoui R et al (2012) Contrasted resistance of stone-dwelling Geodermatophilaceae species to stresses known to give rise to reactive oxygen species. FEMS Microbiol Ecol 80:566–577
Huss VA, Festl H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184–192. doi:10.1016/S0723-2020(83)80048-4
Ivanova N, Sikorski J, Jando M et al (2010) Complete genome sequence of Geodermatophilus obscurus type strain (G-20T). Stand Genomic Sci 2(2):158
Jaouani A, Neifar M, Hamza A et al (2012) Purification and characterization of a highly thermostable esterase from the actinobacterium Geodermatophilus obscurus strain G20. J Basic Microbiol 52:653–660
Jin L, Lee H-G, Kim H-S et al (2013) Geodermatophilus soli sp. nov. and Geodermatophilus terrae sp. nov., two actinobacteria isolated from grass soil. Int J Syst Evol Microbiol 63:2625–2629
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kroppenstedt RM (1982) Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger as stationary phases. J Liq Chromatogr 5:2359–2367
Lechevalier MP, Lechevalier H (1970) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol 20:435–443
Luedemann GM (1968) Geodermatophilus, a new genus of the Dermatophilaceae (Actinomycetales). J Bacteriol 96:1848–1858
Meier-Kolthoff JP, Göker M, Spröer C, Klenk H-P (2013) When should a DDH experiment be mandatory in microbial taxonomy? Arch Microbiol 195:413–418
Mevs U, Stackebrandt E, Schumann P et al (2000) Modestobacter multiseptatus gen. nov., sp. nov., a budding actinomycete from soils of the Asgard Range (Transantarctic Mountains). Int J Syst Evol Microbiol 50(1):337–346
Minnikin DE, O’Donnell AG, Goodfellow M et al (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Montero-Calasanz MC, Göker M, Pötter G et al (2012) Geodermatophilus arenarius sp. nov., a xerophilic actinomycete isolated from Saharan desert sand in Chad. Extremophiles 16:903–909
Montero-Calasanz MC, Göker M, Rohde M et al (2013a) Geodermatophilus siccatus sp. nov., isolated from arid sand of the Saharan desert in Chad. Antonie Van Leeuwenhoek 103:449–456
Montero-Calasanz MC, Göker M, Broughton WJ et al (2013b) Geodermatophilus tzadiensis sp. nov., a UV radiation-resistant bacterium isolated from sand of the Saharan desert. Syst Appl Microbiol 36:177–182
Montero-Calasanz MC, Göker M, Pötter G et al (2013c) Geodermatophilus normandii sp. nov., isolated from Saharan desert sand. Int J Syst Evol Microbiol 63:3437–3443
Montero-Calasanz MC, Göker M, Pötter G et al (2013d) Geodermatophilus saharensis sp. nov., isolated from sand of the Saharan desert in Chad. Arch Microbiol 195:153–159
Montero-Calasanz MC, Göker M, Pötter G et al (2013e) Geodermatophilus africanus sp. nov., a halotolerant actinomycete isolated from Saharan desert sand. Antonie Van Leeuwenhoek 104:207–216
Montero-Calasanz MC, Göker M, Pötter G et al (2013f) Geodermatophilus telluris sp. nov., an actinomycete isolated from Saharan desert sand. Int J Syst Evol Microbiol 63:2254–2259
Montero-Calasanz MC, Göker M, Rohde M et al (2014a) Description of Geodermatophilus amargosae sp. nov., to accommodate the not validly named Geodermatophilus obscurus subsp. amargosae (Luedemann, 1968). Curr Microbiol 68:365–371
Montero-Calasanz MC, Hofner B, Göker M et al (2014b) Geodermatophilus poikilotrophi sp. nov.: a multitolerant actinomycete isolated from dolomitic marble. Biomed Res Int 2014:914767
Nie G-X, Ming H, Li S et al (2012) Geodermatophilus nigrescens sp. nov., isolated from a dry-hot valley. Antonie Van Leeuwenhoek 101:811–817
Normand P (2006) Geodermatophilaceae fam. nov., a formal description. Int J Syst Evol Microbiol 56:2277–2278
Normand P, Benson D (2012) The Actinobacteria. Bergey’s Manual of Systematic Bacteriology, vol 5, 2nd edn. Springer Science & Business Media, Dordrecht, pp 528–530
Normand P, Daffonchio D, Gtari M (2014) The family Geodermatophilaceae. In: The Prokaryotes. Springer, Berlin, Heidelberg, pp 361–379
Qu J-H, Hui M, Qu J-Y et al (2013) Geodermatophilus taihuensis sp. nov., isolated from the interfacial sediment of a eutrophic lake. Int J Syst Evol Microbiol 63:4108–4112
Rainey FA, Ray K, Ferreira M et al (2005) Extensive diversity of ionizing-radiation-resistant bacteria recovered from Sonoran Desert soil and description of nine new species of the genus Deinococcus obtained from a single soil sample. Appl Environ Microbiol 71:5225–5235
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl 20:16
Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477
Sen A, Daubin V, Abrouk D et al (2014) Phylogeny of the class Actinobacteria revisited in the light of complete genomes. The orders “Frankiales” and Micrococcales should be split into coherent entities: proposal of Frankiales ord. nov., Geodermatophilales ord. nov., Acidothermales ord. nov. an. Int J Syst Evol Microbiol 64:3821–3832
Shirling EB, Gottlieb D (1966) Method for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Tamura K, Stecher G, Peterson D et al (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tindall BJ (1990) A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst Appl Microbiol 13:128–130
Vaas LAI, Sikorski J, Michael V et al (2012) Visualization and curve-parameter estimation strategies for efficient exploration of phenotype microarray kinetics. PLoS ONE 7:e34846
Vaas LAI, Sikorski J, Hofner B et al (2013) opm: an R package for analysing OmniLog(R) phenotype microarray data. Bioinformatics 29:1823–1824
Wayne LG, Brenner DJ, Colwell RR et al (1988) International committee on systematic bacteriology announcement of the report of the ad hoc committee on reconciliation of approaches to bacterial systematics. J Appl Bacteriol 64:283–284
Zhi X-Y, Li W-J, Stackebrandt E (2009) An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int J Syst Evol Microbiol 59:589–608
Acknowledgments
This research was supported by The Ministère de l’Enseignement Supérieur et de la Recherche Scientifique, Tunisia (LR03ES03). Chemotaxonomic work was done at the DSMZ (German Collection of Microorganisms and Cell Cultures) Braunschweig.
Conflict of interest
Authors disclose that there are no conflicts of interest. No research involving human participants and/or animals was performed. No non-financial interests tied directly or indirectly to this research exist that may be important to readers to be disclosed.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10482_2015_461_MOESM1_ESM.ppt
Supplementary Fig. S1. The parameter “Maximum Height” estimated from the respiration curves as measured with the OmniLog phenotyping device and discretized and visualized as heatmap using the opm package. Plates and substrates are rearranged according to their overall similarity (as depicted using the row and column dendrograms). Ochre colour indicates positive reaction; purple colour indicates negative reaction; white colour indicates ambiguous reaction. Letters (A/B) indicate each replicate of experiment. Supplementary material 1 (PPT 160 kb)
10482_2015_461_MOESM2_ESM.ppt
Supplementary Fig. S2. Polar lipids profile of strain BMG801T resolved by two-dimensional TLC and revealed by spraying plate with molydatophosphoric acid. DPG diphosphadidylglycerol, PE phosphatidethanolamine, PC phosphatidylcholine, PI phosphatidylinositol, GL unidentified glycolipid, APL unidentified aminophospholipid. Supplementary material 2 (PPT 285 kb)
Rights and permissions
About this article
Cite this article
Hezbri, K., Ghodhbane-Gtari, F., del Carmen Montero-Calasanz, M. et al. Geodermatophilus aquaeductus sp. nov., isolated from the ruins of Hadrian’s aqueduct. Antonie van Leeuwenhoek 108, 41–50 (2015). https://doi.org/10.1007/s10482-015-0461-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-015-0461-z