Abstract
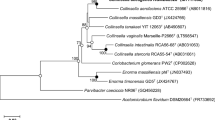
A novel actinobacterial strain, designated CAT-2T, was isolated from human faeces as a bacterium capable of dehydroxylating (+)-catechin derivatives. Strain CAT-2T was found to be strictly anaerobic, Gram-positive, non-motile and non-spore-forming coccobacilli. The major fatty acids were identified as C16:0 DMA (dimethy acetal), C16:0, C14:0, anteiso-C15:0 and iso-C14:0. The three predominant menaquinones were identified as MK-6 (menaquinene-6), MMK-6 (monomethylmenaquinone-6) and DMMK-6 (dimethylmenaquinone-6). The polar lipids were found to be diphosphatidylglycerol, phosphatidylglycerol and four unidentified glycolipid. The DNA G+C content of strain CAT-2T was 68.4 mol%. Phylogenetic analysis based on 16S rRNA gene sequence similarities showed that strain CAT-2T belongs to the genus Gordonibacter, sharing the highest level of sequence homology with Gordonibacter pamelaeae DSM 19378T (97.3 %). Combined phenotypic, chemotaxonomic and phylogenetic characteristics support the conclusion that the strain CAT-2T represents a novel species, for which the name Gordonibacter faecihominis sp. nov. is proposed. The type strain is CAT-2T (= KCTC 15204T = JCM 16058T).



Similar content being viewed by others
References
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1981) Evolutionary trees from DNA sequences, a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies, an approach using the bootstrap. Evolution 39:783–791
Felsenstein J (2009) PHYLIP (phylogeny inference package) version 369. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle, USA
Fitch WM (1971) Toward defining the course of evolution, minimum change for a specific tree topology. Syst Zool 20:406–416
Hall TA (1999) BioEdit, a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Jin JS, Hattori M (2012) Isolation and characterization of a human intestinal bacterium Eggerthella sp. CAT-1 capable of cleaving the C-ring of (+)-catechin and (−)-epicatechin, followed by p-dehydroxylation of the B-ring. Biol Pharm Bull 35:2252–2256
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism, vol 3. Academic Press, New York, pp 21–132
Kohri T, Suzuki M, Nanjo F (2003) Identification of metabolites of (−)-epicatechin gallate and their metabolic fate in the rat. J Agric Food Chem 51:5561–5566
Lányí B (1987) Classical and rapid identification methods for medically important bacteria. Methods Microbiol 19:1–67
Lee KC, Kim KK, Eom MK, Kim MJ, Lee JS (2011) Fontibacillus panacisegetis sp nov, isolated from soil of a ginseng field. Int J Syst Evol Microbiol 61:369–374
Maruo T, Sakamoto M, Ito C, Toda T, Benno Y (2008) Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int J Syst Evol Microbiol 58:1221–1227
Meselhy MR, Nakamura N, Hattori M (1997) Biotransformation of (−)-epicatechin 3-O-gallate by human intestinal bacteria. Chem Pharm Bull 45:888–893
Minnikin DE, Patel PV, Alshamaony L, Goodfellow M (1977) Polar lipid composition in the classification of Nocardia and related bacteria. Int J Syst Bacteriol 27:104–117
Nedashkovskaya OL, Kim SB, Lee MS, Park MS, Lee KH, Lysenko AM, Oh HW, Mikhailov VV, Bae KS (2005) Cyclobacterium amurskyense sp. nov., a novel marine bacterium isolated from sea water. Int J Syst Evol Microbiol 55:2391–2394
Saitou N, Nei M (1987) The neighbour-joining method; a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101 Newark, DE, MIDI Inc
Scheline RR (1970) The metabolism of (+)-catechin to hydroxyphenylvaleric acids by the intestinal microflora. Biochim Biophys Acta 222:228–230
Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477
Shin YK, Lee JS, Chun CO, Kim H-J, Park Y-H (1996) Isoprenoid quinone profiles of Leclercia adecarboxylata KCTC 1036T. J Microbiol Biotechnol 6:68–69
Skerman VBD (1967) A guide to the identification of the genera of bacteria, 2nd edn. Williams Wilkins, Baltimore
Tamaoka J, Komagata K (1984) Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett 25:125–128
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface, flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Wade WG, Downes J, Dymock D, Hiom SJ, Weightman AJ, Dewhirst FE, Paster BJ, Tzellas N, Coleman B (1999) The family Coriobacteriaceae: reclassification of Eubacterium exiguum (Poco et al. 1996) and Peptostreptococcus heliotrinreducens (Lanigan 1976) as Slackia exigua gen. nov., comb. nov. and Slackia heliotrinireducens gen. nov., comb. nov., and Eubacterium lentum (Prevot 1938) as Eggerthella lenta gen. nov., comb. nov. Int J Syst Bacteriol 49:595–600
Wang LQ, Meselhy MR, Li Y, Nakamura N, Min BS, Qin GW, Hattori M (2001) The heterocyclic ring fission and dehydroxylation of catechins and related compounds by Eubacterium sp. strain SDG-2, a human intestinal bacterium. Chem Pharm Bull 49:1640–1643
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE et al (1987) International Committee on Systematic Bacteriology Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Würdemann D, Tindall BJ, Pukall R, Lünsdorf H, Strömpl C, Namuth T, Nahrstedt H, Wos-Oxley M, Ott S, Schreiber S, Timmis KN, Oxley AP (2009) Gordonibacter pamelaeae gen. nov., sp. nov., a new member of the Coriobacteriaceae isolated from a patient with Crohn’s disease, and reclassification of Eggerthella hongkongensis Lau et al. 2006 as Paraeggerthella hongkongensis gen. nov., comb. nov. Int J Syst Evol Microbiol 59:1405–1415
Acknowledgments
This work was supported by the Global R&D Center (GRDC) through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (MSIFP) of the Republic of Korea, by the Bio and Medical Technology Development Program of the NRF funded by the MSIFP of the Republic of Korea, by the KRIBB Research Initiative Program and by the research funds of Chonbuk National University in 2013. I thank Professor J. P. Euzeby for his advice in naming the novel strain.
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of Gordonibacter faecihominis CAT-2T is KF785806.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jin, JS., Lee, K.C., Park, IS. et al. Gordonibacter faecihominis sp. nov., isolated from human faeces. Antonie van Leeuwenhoek 106, 439–447 (2014). https://doi.org/10.1007/s10482-014-0212-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-014-0212-6