Abstract
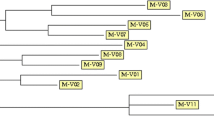
India experienced two plague outbreaks in Gujarat and Maharastra during 1994 and then in the Shimla district of Himachal Pradesh during 2002. Yersinia pestis strains recovered from rodents and pneumonic patients during the 1994 outbreaks, pneumonic patients from the 2002 Shimla outbreak and rodents trapped on the Deccan Plateau during a surveillance activity carried out in 1998 were characterized by MLVA, ERIC-PCR and ERIC-BOX-PCR. MLVA genotyping of Indian Y. pestis strains revealed strains of 2 Orientalis, 1 Mediaevalis and 1 Antiqua genotypes distributed in three distinct branches corresponding to their biovar. The Orientalis genotype strains recovered from the 1994 outbreaks and 1998 surveillance activity clustered in one branch while the Antiqua biovar strains from the Shimla outbreak and the Mediaevalis strain recovered from a rodent trapped on the Deccan Plateau region during surveillance formed the other branches. The Orientalis Y. pestis strains recovered from rodents and patients from the 1994 plague outbreaks exhibited similar MLVA, ERIC-PCR and ERIC-BOX-PCR profiles and these were closely related to the Orientalis strains recovered from the rodents trapped on the Deccan Plateau. These data provide evidence for the possible linkage between the Y. pestis strains resident in the endemic region and those that were associated with the 1994 plague outbreaks. Mediaevalis and Antiqua biovars also were recovered from the environmental reservoir on the Deccan Plateau and from the pneumonic patients of 2002 plague outbreak. Therefore, as in Central Asian and African regions, Antiqua and Mediaevalis biovars seem to be well established in the Indian subcontinent as well. ERIC-PCR DNA fingerprinting delineated genotypes similar to those defined by MLVA. Thus ERIC-PCR appears to have the potential to be used as a molecular marker in the molecular epidemiological investigations of plague.




Similar content being viewed by others
References
Achtman M, Morelli G, Zhu P et al (2004) Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci USA 101:17837–17842. doi:10.1073/pnas.0408026101
Anisimov AP, Lindler LE, Pier GB (2004) Intraspecific diversity of Yersinia pestis. Clin Microbiol Rev 17:434–464. doi:10.1128/CMR.17.2.434-464.2004
Batra HV, Tuteja U, Agarwal GS (1996) Isolation and identification of Yersinia pestis responsible for the recent plague outbreaks in India. Curr Sci 71:787–791
Chain PS, Hu P, Malfatti SA, Radnedge L, Larimer F, Vergez LM, Worsham P, Chu MC, Andersen GL (2006) Complete genome sequence of Yersinia pestis strains Antiqua and Nepal516: evidence of gene reduction in an emerging pathogen. J Bacteriol 188:4453–4463. doi:10.1128/JB.00124-06
Cui Y, Li Y, Gorgé O et al (2008) Insight into microevolution of Yersinia pestis by clustered regularly interspaced short palindromic repeats. PLoS ONE 9;3(7):e2652
De Gregorio E, Silvestro G, Petrillo M, Carlomagno MS, Di Nocera PP (2005) Enterobacterial repetitive intergenic consensus sequence repeats in yersiniae: genomic organization and functional properties. J Bacteriol 187:7945–7954. doi:10.1128/JB.187.23.7945-7954.2005
Drancourt M, Roux V, Dang LV, Tran-Hung L, Castex D, Chenal-Francisque V, Ogata H, Fournier PE, Crubézy E, Raoult D (2004) Genotyping, orientalis-like Yersinia pestis, and plague pandemics. Emerg Infect Dis 10:1585–1592
Dutt AK, Akhtar R, McVeigh M (2006) Review Surat plague of 1994 re-examined. Southeast Asian J Trop Med Public Health 37:755–760
Gage KL, Kosoy MY (2005) Natural history of plague: perspectives from more than a century of research. Annu Rev Entomol 50:505–528. doi:10.1146/annurev.ento.50.071803.130337
Gupta ML, Sharma A (2007) Pneumonic plague, Northern India, 2002. Emerg Infect Dis 13:664–666
Keim P, Price LB, Klevytska AM, Smith KL, Schupp JM, Okinaka R, Jackson PJ, Hugh-Jones ME (2000) Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J Bacteriol 182:2928–2936. doi:10.1128/JB.182.10.2928-2936.2000
Kim W, Song MO, Song W, Kim KJ, Chung SI, Choi CS, Park YH (2003) Comparison of 16S rDNA analysis and rep-PCR genomic fingerprinting for molecular identification of Yersinia pseudotuberculosis. Antonie Van Leeuwenhoek 83:125–133. doi:10.1023/A:1023301924932
Le Flèche P, Hauck Y, Onteniente L, Prieur A, Denoeud F, Ramisse V, Sylvestre P, Benson G, Ramisse F, Vergnaud G (2001) A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol 1:2. doi:10.1186/1471-2180-1-2
Le Flèche P, Fabre M, Denoeud F, Koeck JL, Vergnaud G (2002) High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol 2:37. doi:10.1186/1471-2180-2-37
Lowell JL, Wagner DM, Atshabar B, Antolin MF, Vogler AJ, Keim P, Chu MC, Gage KL (2005) Identifying sources of human exposure to plague. J Clin Microbiol 43:650–656. doi:10.1128/JCM.43.2.650-656.2005
Motin VL, Georgescu AM, Elliot JM et al (2002) Genetic variability of Yersinia pestis isolates as predicted by IS100 genotyping and analysis of structural genes encoding glycerol-3-phosphate dehydrogenase (glpD). J Bacteriol 184:1019–1027. doi:10.1128/jb.184.4.1019-1027.2002
Perry RD, Fetherston JD (1997) Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev 10:35–66
Pourcel C, André-Mazeaud F, Neubauer H, Ramisse F, Vergnaud G (2004) Tandem repeats analysis for the high-resolution phylogenetic analysis of Yersinia pestis. BMC Microbiol 4:22. doi:10.1186/1471-2180-4-22
Sachdeva P, Virdi JS (2004) Repetitive elements sequence (REP/ERIC)-PCR based genotyping of clinical and environmental strains of Yersinia enterocolitica biotype 1A reveal existence of limited number of clonal groups. FEMS Microbiol Lett 240:193–201. doi:10.1016/j.femsle.2004.09.029
Shivaji S, Bhanu NV, Aggarwal RK (2000) Identification of Yersinia pestis as the causative organism of plague in India as determined by 16S rDNA sequencing and RAPD-based genomic fingerprinting. FEMS Microbiol Lett 189:247–252. doi:10.1111/j.1574-6968.2000.tb09238.x
Tacão M, Alves A, Saavedra MJ, Correia A (2005) BOX-PCR is an adequate tool for typing Aeromonas spp. Antonie Van Leeuwenhoek 88:173–179. doi:10.1007/s10482-005-3450-9
Torrea G, Chenal-Francisque V, Leclercq A, Carniel E (2006) Efficient tracing of global isolates of Yersinia pestis by restriction fragment length polymorphism analysis using three insertion sequences as probes. J Clin Microbiol 44:2084–2092. doi:10.1128/JCM.02618-05
van Belkum A, Sluijuter M, de Groot R, Verbrugh H, Hermans PW (1996) Novel BOX repeat PCR assay for high-resolution typing of Streptococcus pneumoniae strains. J Clin Microbiol 34:1176–1179
Versalovic J, Koeuth T, Lupski JR (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 9:6823–6831. doi:10.1093/nar/19.24.6823
Wren BW (2003) The yersiniae—a model genus to study the rapid evolution of bacterial pathogens. Nat Rev Microbiol 1:55–64. doi:10.1038/nrmicro730
Zhou D, Han Y, Song Y et al (2004) DNA microarray analysis of genome dynamics in Yersinia pestis: insights into bacterial genome microevolution and niche adaptation. J Bacteriol 186:5138–5146. doi:10.1128/JB.186.15.5138-5146.2004
Acknowledgment
The authors are thankful to Director, DRDE, Gwalior and Director, DFRL, Mysore for providing us the facilities and support to take up this study. Meticulous reviewing of the manuscript by Dr. J. Pappachan is thankfully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kingston, J.J., Tuteja, U., Kapil, M. et al. Genotyping of Indian Yersinia pestis strains by MLVA and repetitive DNA sequence based PCRs. Antonie van Leeuwenhoek 96, 303–312 (2009). https://doi.org/10.1007/s10482-009-9347-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-009-9347-2