Abstract
Damage of the endothelial glycocalyx (eGC) plays a central role in the development of vascular hyperpermeability and organ damage during systemic inflammation. However, the specific signalling pathways for eGC damage remain poorly defined. Aim of this study was to combine sublingual video-microscopy, plasma proteomics and live cell imaging to uncover further pathways of eGC damage in patients with coronavirus disease 2019 (COVID-19) or bacterial sepsis. This secondary analysis of the prospective multicenter MICROCODE study included 22 patients with COVID-19 and 43 patients with bacterial sepsis admitted to intermediate or intensive care units and 10 healthy controls. Interleukin-6 (IL-6) was strongly associated with damaged eGC and correlated both with eGC dimensions (rs=0.36, p = 0.0015) and circulating eGC biomarkers. In vitro, IL-6 reduced eGC height and coverage, which was inhibited by blocking IL-6 signalling with the anti-IL-6 receptor antibody tocilizumab or the Janus kinase inhibitor tofacitinib. Exposure of endothelial cells to 5% serum from COVID-19 or sepsis patients resulted in a significant decrease in eGC height, which was attenuated by co-incubation with tocilizumab. In an external COVID-19 cohort of 219 patients from Massachusetts General Hospital, a previously identified proteomic eGC signature correlated with IL-6 (rs=-0.58, p < 0.0001) and predicted the combined endpoint of 28-day mortality and/or intubation (ROC-AUC: 0.86 [95% CI: 0.81–0.91], p < 0.001). The data suggest that IL-6 may significantly drive eGC damage in COVID-19 and bacterial sepsis. Our findings provide valuable insights into pathomechanisms of vascular dysfunction during systemic inflammation and highlight the need for further in vivo studies.
Similar content being viewed by others

Introduction
The vascular endothelium is lined by a gel-like matrix of highly sulphated glycosaminoglycans (sGAGs) attached to core proteoglycans, the so-called endothelial glycocalyx (eGC). This fragile structure shields the endothelium from pathogenic insults and plays a key role in maintaining microcirculatory homeostasis. Specifically, the eGC acts as a negatively charged “firewall” to reduce leukocyte-endothelial interactions [1,2,3,4]. Its carbohydrate-rich matrix provides resistance to water permeability and contributes to the proportion of albumin molecules “reflected” back into plasma by the vessel wall [5, 6]. In addition, the glycocalyx contributes to the regulation of the redox state and is critically involved in mediating shear-induced nitric oxide release and physiologic anticoagulation [2, 7, 8]. Inflammation-induced eGC dysfunction leads to vascular hyperpermeability resulting in oedema formation and organ damage in critically ill and particularly septic patients [9, 10].
The ultimate shared pathway of eGC damage, particularly in bacterial sepsis and COVID-19, seems to involve the activation and release of heparanase (HPSE), a heparan sulphate (HS)-degrading enzyme that is unique to mammals and breaks down HS chains from HS proteoglycans found in the glycocalyx [9,10,11]. However, upstream of HPSE upregulation and release, only a few specific signalling pathways have been identified, all of which are pathophysiologically relevant in sepsis. These include the angiopoietin-1/Tie2 ligand receptor system, tumor necrosis factor (TNF)-alpha/TNF receptor signalling and toll-like receptor (TLR)-2 and − 4 signalling [2, 3, 12,13,14].
In this study, we employed sublingual video-microscopy imaging and plasma proteomics to uncover further pathways of eGC damage. The most promising mediator candidate – Interleukin-6 (IL-6) – was subsequently tested for causality by atomic force microscopy in an established endothelial cell (EC) culture system.
Materials and methods
Study design and study population
The clinical part of this study is a secondary analysis of a previous multicenter, prospective, observational, cross-sectional study conducted from May to June 2020 at the University Hospital Münster and three local academic teaching hospitals [15]. The main finding of this study was that the microvascular and proteome signatures of both COVID-19 and bacterial sepsis were very similar and showed almost the same changes compared to healthy controls. We therefore pooled COVID-19 and bacterial sepsis patients in some exploratory analyses in the current study.
After written informed consent was obtained, adult hospitalized patients requiring intensive care (ICU-critical disease) or intermediate care (IMC-moderate/severe disease) because of COVID-19 infection or confirmed bacterial sepsis (sepsis-3 definition) [16] were prospectively enrolled in a non-consecutive manner. Sublingual video-microscopy was performed at the same time as blood sampling. Plasma samples were collected, centrifuged, and stored at -80 °C until analysis. Exclusion criteria were pregnancy or local inflammation of the oral mucosa. None of the patients received therapy specifically targeting IL-6. Ten apparently healthy, randomly selected age-matched volunteers served as controls. Three randomly selected serum samples with high IL-6 levels (from the third IL-6 tertile) of each COVID-19 and sepsis were used for in vitro experiments. This study was approved by the local ethics committee (amendments of 2016–073-f-S) and was conducted in accordance with the Declaration of Helsinki.
In vivo assessment of sublingual microcirculation and glycocalyx dimensions
A sidestream dark field camera (CapiScope HVCS, KK Technology, Honiton, UK) coupled with GlycoCheck™ software (Microvascular Health Solutions Inc., Alpine, UT, USA) was used to visualize passing red blood cell (RBC) flow in the sublingual microvasculature (microvessel diameter 4–25 μm) at the bedside as previously described in detail [17, 18]. Based on the RBC dynamics in the valid vessel segments, the software calculates the following variables, which have been successfully validated in the past [12, 17,18,19]:
Perfused Boundary Region (PBR, in µm) expresses the dynamic lateral movement of RBCs into the permeable part of the endothelial glycocalyx layer, an inverse parameter of the endothelial glycocalyx thickness. The higher the PBR values, the thinner the glycocalyx (Supplemental Fig. 1).
Capillary density (in 10− 2mm/mm2) was defined as the vascular density of vessels with a diameter ≤ the diameter of a single erythrocyte (diameter ~ 7–8 μm [20]; capillary density diameter 4–7 μm).
RBC velocity (in µm/sec) can be automatically determined in individual vessel segments by cross-correlating RBC longitudinal intensity profiles between frames of recorded videos.
Targeted plasma proteomics and circulating glycocalyx markers
The “Inflammation1” and “Cardiovascular2” proteomic panels from Olink (Sweden) each contained 92 proteins. Seven proteins (including IL-6) were common to both panels. A total of 184 proteins in 76 samples (COVID-19, bacterial sepsis, healthy) were measured in one batch to avoid technical variation. In brief, two specific oligonucleotide-labelled antibodies per protein (‘probes’) were used in the Olink proximity extension assay. When the two probes were in close proximity to each other, a new PCR target sequence was formed via a proximity dependent DNA polymerisation event. The resulting sequence was then detected and quantified by standard real-time quantitative PCR as previously reported [21]. Measurements were performed in triplicates. Results were expressed in arbitrary units. The proteins included in each panel, measurement details and validation data are available online (www.olink.com/downloads).
Plasma levels of the glycocalyx core protein syndecan-1 (Diaclone, Besançon, France) and hyaluronic acid (Echelon Biosciences Inc., Salt Lake City, UT, USA) were measured using commercially available enzyme-linked immunosorbent assay kits according to the manufacturer’s instructions.
External validation set ─ Massachusetts General Hospital COVID-19 cohort
Some findings were validated in a public database of adult COVID-19 patients admitted to the Massachusetts General Hospital (MGH, Boston, Massachusetts, USA, https://www.olink.com/mgh-covid-study), which annotates proteomic and outcome data [22]. Inclusion criteria were clinical concern for COVID-19 on admission to the emergency department and acute respiratory distress with at least one of the following: respiratory rate ≥ 22 breaths/minute; oxygen saturation ≤ 92% on room air; need for supplemental oxygen; positive pressure ventilation. The primary endpoint of the study was a composite endpoint of 28-day mortality and/or intubation, and a total of 219 blood samples collected on day 3 were analyzed in relation to the primary endpoint.
Cell culture and reagents
Cells of the human umbilical vein endothelial cell (HUVEC) line EA.hy926 were grown in DMEM (Gibco™; Cat# 52100047) supplemented with 10% fetal calf serum (FCS, SigmaAldrich) and 1% penicillin/streptomycin (Biochrom; Cat# A2212) at 37 °C in a 5% CO2 enriched environment for a minimum of 3 days until reaching confluence.
Experiments were conducted in (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (HEPES) buffer (140 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 5 mM glucose, 10 mM HEPES) supplemented with 1% FCS and incubation times of 60 min if not otherwise stated. IL-6 (#200-06) and sIL-6R (#200-06RC) were purchased from PeproTech (Thermo Fisher Scientific, Germany), Heparin (#H3149) and Tofacitinibcitrate (#PZ0017) from Sigma-Aldrich (Germany), and Tocilizumab (EU: RoActemra®, US: Actemra®) from Roche (Germany).
1,9-Dimethylmethylene blue assay
To measure the amount of sGAGs in the supernatant samples, the 1,9-Dimethylmethylene Blue (DMMB) assay was performed as previously described [23, 24]. Briefly, after incubation, the cell supernatant was collected and concentrated by centrifugation in Microcon-10 kDa centrifugal filter units (Merck; Cat# MRCPRT010) for 20 min at 14,000 x g. 50 µl of the concentrate were transferred in duplicates to a 96-well plate (Greiner bio-one; Cat# 655,101). 200 µl of the DMMB buffer was added per well and the absorbance was immediately read at 525 and 590 nm (Tecan, Infinite M200). A standard curve with chondroitin sulfate-4 served as a control. Data were presented standardized to the corresponding control condition.
Atomic force microscopy
Quantification of eGC thickness in vitro was performed by atomic force microscopy (AFM) nanoindentation technique using a Nanoscope V multimode AFM (Veeco, Mannheim, Germany) as previously described in detail [3, 12]. The measurement was performed in a liquid chamber at 37 °C in HEPES buffer supplemented with 1% FCS. Periodic indentation and deflection of the triangular cantilever with a mounted 10 μm spherical tip (Novascan Technologies, Boone, North Carolina, United States) - spring constant 10 pN/nm – was detected by a laser beam. The eGC thickness of the indented area was calculated from the resulting force versus distance curve. By measuring at least 24 cells per group/condition, AFM can detect significant differences of at least 15% between three groups with a power of 95% power (G*Power 3.1). Data were presented standardized to the corresponding control condition.
Immunofluorescence
Heparan sulphate (HS) staining was performed essentially as described in previous studies [11, 14, 25]. Cells were fixed with 2% paraformaldehyde plus 0.1% glutaraldehyde followed by overnight incubation with primary antibody (Amsbio, Ab heparan sulfate, Cat# 370255-1) and secondary antibody (Jackson ImmunoResearch Labs, Alexa Fluor 488 goat anti-mouse IgG antibody, Cat# 115-545-146) and 4,6-diamidino-2-phenylindole dihydrochloride (DAPI). Images were captured with a Leica DMI 6000B-CS/TCS SP8 laser confocal microscope (objective: HC PL APO CS2 63 × /1.40 oil; Leica, Wetzlar, Germany) and analyzed using LasX software (Leica, Wetzlar, Germany) and ImageJ software (version 1.51p 22, National Institutes of Health, United States).
Statistics
Data were presented as indicated with median ± interquartile range (IQR) or mean ± standard error of mean (SEM), unless otherwise noted. Differences between two groups were calculated by Mann-Whitney U test or chi-square, as appropriate. Comparisons between ≥ 3 groups were performed with Kruskal-Wallis test with Dunn’s post-hoc test. The Spearman correlation coefficient (rs) was used to assess correlations between variables. For the AFM and DMMB experiments, nested ANOVA with Tukey’s post-hoc test was used to account for the number of observations per experiment and the number of experiments.
Analysis and visualization of protein expression data was performed using the R software package (version 4.2.1) [26]. After quality control, one Covid-19 ICU outlier sample was excluded (very low median expression level compared to all other samples). For identification of differentially expressed proteins (DEPs), the Limma package (version 3.52.4 [27, 28]), was used with the design: model.matrix(~ 0 + group). For the contrast of infected versus healthy controls, groups were: all infected (bacterial sepsis and COVID-19 combined, n = 65) and healthy controls. For the other comparisons, groups were: healthy controls, COVID-19 ICU, bacterial sepsis ICU. DEPs were identified based on an adjusted p-value of < 0.05 and exhibiting more than a 1.5-fold (log2 = 0.5849625) difference in expression levels. Multiple testing adjusted p-value were calculated according to Benjamini and Hochberg [29]. Volcano plots were generated with the package EnhancedVolcano, version 1.14.0 [30].
All tests were two-tailed and significance was accepted at p < 0.05. GraphPad Prism version 9 (GraphPad Prism Software Inc, San Diego, California, USA) and SPSS 29 (IBM, Armonk, New York, NY, USA) were used for further data analysis and figure preparation.
Results
Our cohort consisted of 43 patients with bacterial sepsis and 22 with COVID-19. There were no significant differences between median [IQR] age (68 [57–79] vs. 63 [53–76] years, p = 0.12), sex (p = 0.14), or disease severity (Sequential Organ Failure Assessment (SOFA) score 9 [4-12] vs. 6 [2-12], p = 0.22) in the two groups (Table 1). ICU patients had a higher SOFA score than IMC patients (10 [6-13] vs. 2 [0.5-3], p < 0.0001) and were ventilated in 66,7% of cases. IMC patients were more likely to be female (47.1% vs. 18.8%, p = 0.023), but did not differ from ICU patients in terms of age (64 [55-79.5] vs. 64.5 [56.3–76.5] years, p = 0.69) (Supplemental Table 1).
Proteome analysis identifies IL-6 as potential mediator of eGC damage in inflammation
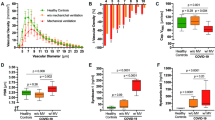
ICU patients with either sepsis or COVID-19 had significantly higher PBR4 − 25 μm values (i.e., thinner endothelial glycocalyx) than healthy controls or IMC patients, respectively (Fig. 1A, Supplemental Table 1). A pooled analysis including COVID-19 and bacterial sepsis patients showed that PBR values correlated with the SOFA score (rs = 0.28, p = 0.016), indicating that disease severity correlates with eGC damage.
To identify potential mediators of glycocalyx damage, we performed differentially expressed protein (DEP) analysis after dividing participants into those with intact vs. damaged eGC (Supplemental Fig. 2). When compared to participants with intact eGC, there were 31 up- and 2 downregulated DEPs in participants with damaged eGC. Of these, IL-6 showed by far the largest log2 fold change of all 177 proteins (Fig. 1B). This finding was consistently reproducible and even more pronounced when classification was based on disease entity rather than eGC, suggesting that IL-6 upregulation was relevant in both COVID-19 and sepsis (Supplemental Fig. 3).
Median IL-6 levels measured at routine laboratory were significantly higher in patients with damaged eGC than in those with intact eGC (131 [69–826] vs. 30 [2–90] ng/ml, p = 0.0003; Supplemental Table 2). Furthermore, IL-6 levels correlated with the PBR (rs=0.36, p = 0.0015) and circulating biomarkers of eGC damage (syndecan-1: rs = 0.41, p = 0.0004; hyaluronan: rs = 0.58, p < 0.0001). When visualized by overall IL-6 tertiles, PBR, syndecan-1 and hyaluronan showed a steady increase (Fig. 1C-E). Consistent with previous work, showing an uncoupling of PBR (eGC integrity) and microvascular perfusion [17, 19], IL-6 levels were only weakly associated with capillary density (rs = -0.27, p = 0.018) (Supplemental Table 2, Supplemental Fig. 4).
Proteome analysis identifies IL-6 as potential mediator of eGC damage in COVID-19 and bacterial sepsis. (A) Box plots (median ± IQR) of perfused boundary region (PBR) values from sublingual video-microscopy in healthy controls, intermediate care (IMC) and intensive care (ICU) patients. (B) Volcano plot showing log2-fold changes and adjusted p-values of differentially regulated proteins (DRPs) for low vs. high PBR groups (using a cut-off of 2.25 μm). As IL-6 is included in both proteomic panels, it appears twice. Box plots of (C) PBR, (D) circulating syndecan-1 (available in 72 subjects) and (E) circulating hyaluronan (available in 72 subjects) in healthy controls vs. patients (IL-6-derived tertiles, first tertile = lowest IL-6 levels). Individual values are shown as green dots = healthy controls; blue dots = COVID-19; pink dots = sepsis. Significance was tested with Mann-Whitney (also see Supplemental Table 1) or Kruskal-Wallis and Dunn’s post-hoc test against healthy controls. *=p < 0.05, **=p < 0.01
IL-6 signalling causes eGC damage in vitro
In classical IL-6 signalling, IL-6 binds to the membrane-bound IL-6 receptor α subunit (hereafter mIL-6R) and the glycoprotein 130 (gp130) signal-transducing subunit. In contrast, in IL-6 trans-signalling, complexes of IL-6 and the soluble form of the IL-6 receptor (sIL-6R) signal via (ubiquitously expressed) membrane-bound gp130. HUVECs express both receptors, gp 130 and small amounts of mIL-6R [31, 32].
Incubation of ECs with low to supra-physiological concentrations (0.1–10 ng/ml) of IL-6 alone resulted in a weak and dose-independent increase in sGAG content in the endothelial cell supernatant (data not shown), suggesting that damage to the eGC by classical signalling mechanisms is negligible. However, the addition of a physiologic concentration of sIL-6R (50 ng/ml) to simulate trans-signalling showed a doubling of the amount of sGAG in the supernatant in response to an intermediate concentration of IL-6, consistent with a more potent effect of trans-signalling (Fig. 2A). Accordingly, 2D images and 3D reconstructions of immunofluorescence-stained heparan sulphate, the major GAG of the eGC, showed that the intensity and coverage of the EC surface is reduced after incubation with IL-6/sIL-6R (Fig. 2B, C). As systemic trans-signalling dominates in acute inflammatory responses such as sepsis and COVID-19 [33], all further in vitro experiments were performed with the combination of IL-6 (1 ng/ml) and sIL-6R (50 ng/ml).
We then used the much more accurate nano-indentation AFM method to investigate clinically available inhibitors of IL-6 signalling in vitro. Co-incubation with the humanized monoclonal antibody tocilizumab (100 µg/ml), an antibody against the IL-6R that prevents IL-6 from binding to the IL-6R, completely prevented IL-6-induced glycocalyx damage on ECs (Fig. 2D). Similar results were obtained by blocking Janus kinases (JAKs), which act downstream of gp130, with tofacitinib (10 µM, pre-incubation for 24 h) (Fig. 2E).
We have previously shown that in sepsis and COVID-19, the final common pathway of eGC damage appears to be the activation and release of the heparan sulphate-degrading enzyme heparanase [11, 12]. The addition of heparin - a potent heparanase inhibitor - also protected eGC from IL-6-induced damage, suggesting that IL-6 signalling ultimately acts to regulate heparanase (Fig. 2F).
IL-6 trans-signalling causes eGC damage in vitro. (A) The amount of sulphated glycosaminoglycans (sGAGs) in the supernatant of endothelial cells was measured using the 1,9-dimethylmethylene blue (DMMB) assay. Cells were incubated for 60 min with the indicated concentrations of IL-6 ± sIL-6R, TNFα or LPS supplemented with CD14 (10 ng/ml) and LBP (100 ng/ml). Data are expressed as mean ± SEM of duplicates, n = 3–9. (B + C) Immunofluorescence imaging showing the distribution and coverage of heparan sulphate (green) staining on the cell layer in 3D reconstruction (B) derived from 2D images (C). Nuclei were stained with DAPI (blue). Scale bar 10 μm. Incubation was performed under the same conditions as described in (A) with IL-6 (1 ng/ml) + sIL-6R (50 ng/ml). Incubation in Hepes buffer supplemented with 1% fetal calf serum was used as a control. (D-F) Nanoindentation experiments with atomic force microscopy (AFM) showing changes of eGC height on living endothelial cells after 60 min incubation with sIL-6R + IL-6 ± tocilizumab, tofacitinib or heparin. Incubation in Hepes buffer supplemented with 1% fetal calf serum was used as control. All conditions were pre-incubated for 24 h with vehicle or in case of tofacitinib treatment with tofacitinib. Data are presented as mean ± SEM, each point represents the average of ≥ 4 indentations per cell with a minimum of 8 cells per experiment, n = 3-4. Significance was tested by nested ANOVA followed by Tukey’s or Dunnett’s post-hoc test. *=p < 0.05, **=p < 0.01
Tocilizumab protects from serum-induced eGC damage in bacterial sepsis and COVID-19
To simulate more realistic inflammatory conditions in the context of sepsis and COVID-19, three randomly selected serum samples from the third tertile (highest IL-6 concentrations) of each COVID-19 and sepsis were pooled and used for the following in vitro experiments. Mean [± SEM] IL-6 concentrations in the pooled sepsis subgroup were higher than in the COVID-19 subgroup (2859 [± 1275] vs. 636 [± 476] ng/ml). Incubation of ECs with pooled serum (5% for 60 min) resulted in a significant decrease in eGC in both sepsis and COVID-19. Co-incubation with tocilizumab (100 µg/ml) protected the eGC from COVID-19 serum-induced damage (Fig. 3A). A similar trend was observed with co-incubation of tocilizumab with sepsis serum, although it was not statistically significant (Fig. 3B).
Tocilizumab protects from serum-induced eGC damage in bacterial sepsis and COVID-19. Nanoindentation experiments with atomic force microscopy (AFM) showing changes of eGC height on living endothelial cells after 60 min incubation with pooled serum samples (5%) from (A) COVID-19 and (B) sepsis patients with or without concomitant addition of tocilizumab. Pooled sera from healthy individuals were used as control. Data are presented as mean ± SEM, each point represents the average of ≥ 4 indentations per cell with a minimum of 8 cells per experiment, n = 3–4. Significance was tested by nested ANOVA followed by Tukey’s post-hoc test, *=p < 0.05, **=p < 0.01
Proteome-derived eGCsignature correlates with IL-6 and outcome in external validation set
In the last step, we wanted to validate the mechanistic link between PBR and IL-6. As our study was not designed for outcome analysis, we used the external MGH COVID-19 cohort, which annotates proteome and outcome data. As sublingual microscopy was not performed in the MGH cohort, we used a previously validated proteomic signature that correlates well with eGC thickness as a surrogate (hereafter referred to as eGCsignature) [15]. Patients’ eGCsignature values correlated inversely well with IL-6 levels (rs = -0.58, p < 0.0001) (Fig. 4A). When dichotomized by the median, patients with lower IL-6 levels had a higher eGCsignature, values indicating a healthier eGC (p < 0.0001) (Fig. 4B). For the composite endpoint of 28-day mortality and/or intubation, eGCsignature performed similarly to IL-6 (AUC [95% CI] 0.86 [0.81–0.91], p < 0.001] vs. IL-6 0.88 [0.83–0.92], p < 0.001), further supporting the proposed mechanistic role of IL-6 in eGC damage (Fig. 4C).
Proteome-derived eGCsignature correlates with IL-6 and outcome in external validation set. External validation of a previously identified proteomic signature that correlates well with eGC thickness (hereafter referred to as eGCsignature) in an independent COVID-19 cohort from Massachusetts General Hospital (n = 219). (A) Dot plot analysis showing the correlation between the eGCsignature and IL-6. (B) eGCsignature classified based on median IL-6 levels. (C) Receiver operating characteristic curves showing the predictive ability of eGCsignature and IL-6 to predict the composite endpoint of 28-day mortality and/or intubation. AU arbitrary units. **p < 0.01
Discussion
This study demonstrates that IL-6 and its downstream signalling have a causal role in eGC damage. In vitro pharmacological IL-6 blockade protected against eGC damage induced by sera from bacterial sepsis and COVID-19 patients. Additionally, IL-6 levels correlated with features of eGC impairment and predicted outcomes in an external COVID-19 cohort. These data clearly suggest that IL-6 may be a significant driver of eGC damage during systemic inflammation.
A first hint suggesting a possible link between IL-6 and the eGC came from Ikonomidis et al. who observed a decrease of the PBR (i.e. improvement of the eGC) upon tocilizumab administration in patients with rheumatoid arthritis [34]. In the case of COVID-19, evidence of eGC damage has been found in numerous studies [35,36,37]. However, the pathophysiological pathways that trigger this damage are not yet fully understood.
By establishing a causal role for IL-6 in eGC damage, we add an important piece of the puzzle to the existing literature. Therapeutic strategies targeting IL-6 have been successful in the treatment of severe COVID-19 disease, making this finding even more exciting, and regulatory authorities have recently approved its therapeutic use [38,39,40,41,42]. A recent meta-analysis [43] showed that IL-6 blockade with tocilizumab works best for moderate to severe COVID-19. Contrary to expectations, secondary infections were not increased with the administration of IL-6 receptor antagonists in COVID-19 patients. However, IL-6R blockade increases the risk of bacterial, viral, and opportunistic infections in rheumatoid arthritis [44] and should therefore not be used in bacterial sepsis.
In our cell culture model, blocking not only binding of IL-6 to IL-6R, but also its downstream JAK/STAT pathway with tofacitinib was sufficient to counteract eGC damage. Clinical studies on hospitalized COVID-19 patients have also demonstrated a survival benefit for this class of substances [45,46,47]. However, as none of the clinical trials on anti-IL-6 therapy in COVID-19 used sublingual microscopy to estimate the eGC properties, we are currently unable to estimate the effect of anti-IL-6 treatment on eGC dimensions in vivo. However, due to the widespread systemic vascular involvement in COVID-19, it appears plausible that anti-IL-6 therapy would have reduced damage to the eGC. This hypothesis is further supported by the strong correlation between IL-6 and eGC signature in the validation cohort.
Although it is currently unclear whether IL-6 inhibition has similar benefits in sepsis, a recent Mendelian randomization analysis suggests that IL-6 receptor blockade is associated with lower mortality in 11,643 sepsis patients of the UK Biobank cohort [48]. Barkhausen et al. reported that in a murine polymicrobial sepsis model, pretreatment with a selective inhibitor of IL-6 trans-signalling increased survival from 45 to 100% in a dose-dependent manner [49]. Similarly, specific inhibition of IL-6 trans-signalling completely prevented death in mice with endotoxic shock [50]. Kang et al. also detected improved survival in a murine endotoxic shock model by counteracting vascular injury upon treatment with anti-IL-6R antibody. They could further demonstrate in a murine burn injury model, that short-term IL-6R inhibition preserved eGC integrity in capillaries visualized by electron microscopy [51]. These studies support the results of our in vitro experiments and emphasize the central importance of IL-6 in bacterial sepsis, which is also reflected in its prominent role as an early diagnostic and prognostic biomarker [52,53,54].
However, it is important to note that the preventative effect of tocilizumab on serum-induced eGC damage was somewhat weaker in bacterial sepsis compared to COVID-19. It is unlikely that this is only due to a pure dose effect of IL-6 (which is about four times higher in sepsis), as tocilizumab was administered in a saturating dose. It is more likely that eGC in sepsis is affected by additional and/or more complex mechanisms, which may explain the lack of clinical efficacy of several targets that (at least in theory) should also diminish heparanase release [55]. Further research is required to determine the role of other harmful mediators in the complex cytokine milieu during systemic inflammation. The aim should be to identify the smallest possible set of key mediators or effectors whose blockade can prevent eGC damage in a non-redundant manner. In the case of COVID-19, the singular blockade of IL-6 already appears to be close to achieving this goal.
As this study is primarily hypothesis-generating, it is important to note some limitations. Firstly, the sample size of this cross-sectional study was rather small and was not suitable for the analysis of clinical outcomes. However, the comprehensive dataset combines serum proteomics with intravital microscopy. Secondly, it was not our intention to make a direct comparison between the two entities, but rather to analyze IL-6 in the context of systemic inflammation, using sepsis and COVID-19 as prototypical diseases. Although the entities were initially pooled, the supergroup analyses clearly show that IL-6 appears to play an important role in both. Thirdly, although routine microbiological sampling was performed in all patients, we cannot exclude the possibility of bacterial superinfections in the COVID-19 group, which may have partially influenced IL-6 levels. However, the prevalence of bacterial co-infections in COVID-19 is considered rather low and our COVID-19 cohort had a low median PCT value (0.6 ng/ml), which argues against overt co-infections. Fourthly, measuring the eGC both in vivo and in vitro presents a challenge due to the fragility of this delicate layer. Therefore, we complemented intravital microscopy and atomic force microscopy with additional detection methods, such as immunofluorescence and ELISA measurements. These methods broadly confirmed the microscopy data. However, it is crucial to acknowledge that determining PBR through intravital microscopy and measuring eGC thickness using AFM have limitations, which must be considered when interpreting the results. The AFM method may only detect the denser parts of the eGC. However, a strong correlation between the two methods has been observed multiple times using matched data [12, 19]. A detailed description of the advantages and disadvantages of these and other methods was published recently [4]. Finally, it is possible that randomly selecting and pooling of 3 sera from the upper percentile of each group for in vitro experiments may have introduced bias. However, this approach has been a valid compromise in our previous AFM studies [12, 19].
Conclusion
Our data reveal a novel mechanistic pathway elucidating endothelial glycocalyx damage in inflammatory states such as bacterial sepsis and COVID-19. Further in vivo studies should validate our findings and demonstrate the protective efficacy of potential therapeutic interventions targeting IL-6 signalling on the endothelial glycocalyx. These trials will not only improve our pathomechanistic understanding but also pave the way for targeted interventions.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AU:
-
Arbitrary unit
- AUC:
-
Area under the curve
- COVID-19:
-
Coronavirus disease 2019
- CRP :
-
C-reactive protein
- DMMB:
-
1,9-Dimethylmethylene Blue
- PCT:
-
Procalcitonin
- EC:
-
Endothelial cell
- eGC:
-
Endothelial glycocalyx
- HPSE:
-
Heparanase
- HS:
-
Heparan sulphate
- HUVEC:
-
Human umbilical vein endothelial cell
- ICU:
-
Intensive care unit
- IL-6:
-
Interleukin-6
- IL-6R :
-
Interleukin-6-receptor
- IMC:
-
Intermediate care
- IQR:
-
Interquartile range
- PBR:
-
Perfused boundary region
- RBC:
-
Red blood cell
- SEM:
-
Standard error of the mean
- sGAG:
-
Sulphated glycosaminoglycan
- SOFA:
-
Sequential organ failure assessment
- TLR:
-
Toll-like receptor
- TNF:
-
Tumor necrosis factor
References
Mulivor AW HHL (2002) Role of glycocalyx in leukocyte-endothelial cell adhesion. Am J Physiol Heart Circ Physiol 283(4):H1282–1291
Schmidt EP, Yang Y, Janssen WJ, Gandjeva A, Perez MJ, Barthel L, Zemans RL, Bowman JC, Koyanagi DE, Yunt ZX, Smith LP, Cheng SS, Overdier KH, Thompson KR, Geraci MW, Douglas IS, Pearse DB, Tuder RM (2012) The pulmonary endothelial glycocalyx regulates neutrophil adhesion and lung injury during experimental sepsis. Nat Med 18(8):1217–1223. https://doi.org/10.1038/nm.2843
Wiesinger A, Peters W, Chappell D, Kentrup D, Reuter S, Pavenstadt H, Oberleithner H, Kumpers P (2013) Nanomechanics of the endothelial glycocalyx in experimental sepsis. PLoS ONE 8(11):e80905. https://doi.org/10.1371/journal.pone.0080905
Drost CC, Unger A, Linke WA, Vink H, Kumpers P (2023) The dynamics of the endothelial glycocalyx: a rare snapshot by electron microscopy. Angiogenesis 26(4):487–491. https://doi.org/10.1007/s10456-023-09880-x
Curry FE, Adamson RH (2012) Endothelial glycocalyx: permeability barrier and mechanosensor. Ann Biomed Eng 40(4):828–839. https://doi.org/10.1007/s10439-011-0429-8
Salmon AH, Ferguson JK, Burford JL, Gevorgyan H, Nakano D, Harper SJ, Bates DO, Peti-Peterdi J (2012) Loss of the endothelial glycocalyx links albuminuria and vascular dysfunction. J Am Soc Nephrol 23(8):1339–1350. https://doi.org/10.1681/ASN.2012010017
Alphonsus CS, Rodseth RN (2014) The endothelial glycocalyx: a review of the vascular barrier. Anaesthesia 69(7):777–784. https://doi.org/10.1111/anae.12661
Padberg JS, Wiesinger A, di Marco GS, Reuter S, Grabner A, Kentrup D, Lukasz A, Oberleithner H, Pavenstadt H, Brand M, Kumpers P (2014) Damage of the endothelial glycocalyx in chronic kidney disease. Atherosclerosis 234(2):335–343. https://doi.org/10.1016/j.atherosclerosis.2014.03.016
Uchimido R, Schmidt EP, Shapiro NI (2019) The glycocalyx: a novel diagnostic and therapeutic target in sepsis. Crit Care 23(1):16. https://doi.org/10.1186/s13054-018-2292-6
Drost CC, Rovas A, Kumpers P (2021) Protection and rebuilding of the endothelial glycocalyx in sepsis - science or fiction? Matrix Biol Plus 12:100091. https://doi.org/10.1016/j.mbplus.2021.100091
Drost CC, Rovas A, Osiaevi I, Rauen M, van der Vlag J, Buijsers B, Salmenov R, Lukasz A, Pavenstadt H, Linke WA, Kumpers P (2022) Heparanase is a putative mediator of endothelial glycocalyx damage in COVID-19 - a proof-of-Concept Study. Front Immunol 13:916512. https://doi.org/10.3389/fimmu.2022.916512
Drost CC, Rovas A, Kusche-Vihrog K, Van Slyke P, Kim H, Hoang VC, Maynes JT, Wennmann DO, Pavenstadt H, Linke W, Lukasz A, Hesse B, Kumpers P (2019) Tie2 activation promotes Protection and reconstitution of the endothelial glycocalyx in human Sepsis. Thromb Haemost 119(11):1827–1838. https://doi.org/10.1055/s-0039-1695768
Lei J, Xiang P, Zeng S, Chen L, Zhang L, Yuan Z, Zhang J, Wang T, Yu R, Zhang W, Ibrahim II, Ma L, Yu C (2021) Tetramethylpyrazine alleviates endothelial glycocalyx degradation and promotes glycocalyx restoration via TLR4/NF-kappaB/HPSE1 signaling pathway during inflammation. Front Pharmacol 12:791841. https://doi.org/10.3389/fphar.2021.791841
Hesse B, Rovas A, Buscher K, Kusche-Vihrog K, Brand M, Di Marco GS, Kielstein JT, Pavenstadt H, Linke WA, Nofer JR, Kumpers P, Lukasz A (2020) Symmetric dimethylarginine in dysfunctional high-density lipoprotein mediates endothelial glycocalyx breakdown in chronic kidney disease. Kidney Int 97(3):502–515. https://doi.org/10.1016/j.kint.2019.10.017
Rovas A, Buscher K, Osiaevi I, Drost CC, Sackarnd J, Tepasse PR, Fobker M, Kuhn J, Braune S, Gobel U, Tholking G, Groschel A, Rossaint J, Vink H, Lukasz A, Pavenstadt H, Kumpers P (2022) Microvascular and proteomic signatures overlap in COVID-19 and bacterial sepsis: the MICROCODE study. Angiogenesis 25(4):503–515. https://doi.org/10.1007/s10456-022-09843-8
Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent JL, Angus DC (2016) The Third International Consensus definitions for Sepsis and septic shock (Sepsis-3). JAMA 315(8):801–810. https://doi.org/10.1001/jama.2016.0287
Rovas A, Sackarnd J, Rossaint J, Kampmeier S, Pavenstadt H, Vink H, Kumpers P (2021) Identification of novel sublingual parameters to analyze and diagnose microvascular dysfunction in sepsis: the NOSTRADAMUS study. Crit Care 25(1):112. https://doi.org/10.1186/s13054-021-03520-w
Rovas A, Lukasz AH, Vink H, Urban M, Sackarnd J, Pavenstadt H, Kumpers P (2018) Bedside analysis of the sublingual microvascular glycocalyx in the emergency room and intensive care unit - the GlycoNurse study. Scand J Trauma Resusc Emerg Med 26(1):16. https://doi.org/10.1186/s13049-018-0483-4
Rovas A, Seidel LM, Vink H, Pohlkotter T, Pavenstadt H, Ertmer C, Hessler M, Kumpers P (2019) Association of sublingual microcirculation parameters and endothelial glycocalyx dimensions in resuscitated sepsis. Crit Care 23(1):260. https://doi.org/10.1186/s13054-019-2542-2
Diez-Silva M, Dao M, Han J, Lim CT, Suresh S (2010) Shape and Biomechanical Characteristics of Human Red Blood Cells in Health and Disease. MRS Bull 35(5):382–388. https://doi.org/10.1557/mrs2010.571
Assarsson E, Lundberg M, Holmquist G, Bjorkesten J, Thorsen SB, Ekman D, Eriksson A, Rennel Dickens E, Ohlsson S, Edfeldt G, Andersson AC, Lindstedt P, Stenvang J, Gullberg M, Fredriksson S (2014) Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE 9(4):e95192. https://doi.org/10.1371/journal.pone.0095192
Filbin MR, Mehta A, Schneider AM, Kays KR, Guess JR, Gentili M, Fenyves BG, Charland NC, Gonye ALK, Gushterova I, Khanna HK, LaSalle TJ, Lavin-Parsons KM, Lilley BM, Lodenstein CL, Manakongtreecheep K, Margolin JD, McKaig BN, Rojas-Lopez M, Russo BC, Sharma N, Tantivit J, Thomas MF, Gerszten RE, Heimberg GS, Hoover PJ, Lieb DJ, Lin B, Ngo D, Pelka K, Reyes M, Smillie CS, Waghray A, Wood TE, Zajac AS, Jennings LL, Grundberg I, Bhattacharyya RP, Parry BA, Villani AC, Sade-Feldman M, Hacohen N, Goldberg MB (2021) Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions. Cell Rep Med 2(5):100287. https://doi.org/10.1016/j.xcrm.2021.100287
Schmidt EP, Overdier KH, Sun X, Lin L, Liu X, Yang Y, Ammons LA, Hiller TD, Suflita MA, Yu Y, Chen Y, Zhang F, Cothren Burlew C, Edelstein CL, Douglas IS, Linhardt RJ (2016) Urinary glycosaminoglycans predict outcomes in septic shock and Acute Respiratory Distress Syndrome. Am J Respir Crit Care Med 194(4):439–449. https://doi.org/10.1164/rccm.201511-2281OC
Rovas A, Neumann JK, Drost CC, Vollenberg R, Tholking G, Fobker M, Witzenrath M, Kumpers P, Agamotto Study G, Capnetz Study G (2023) Analysis of urinary glycosaminoglycans to Predict Outcome in COVID-19 and Community-Acquired Pneumonia-A Proof-of-Concept Study. J Clin Med 12(16). https://doi.org/10.3390/jcm12165269
Lukasz A, Hillgruber C, Oberleithner H, Kusche-Vihrog K, Pavenstadt H, Rovas A, Hesse B, Goerge T, Kumpers P (2017) Endothelial glycocalyx breakdown is mediated by angiopoietin-2. Cardiovasc Res 113(6):671–680. https://doi.org/10.1093/cvr/cvx023
R_Core_Team (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria URL http://www.R-project.org/
Smyth GK (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3:Article3. https://doi.org/10.2202/1544-6115.1027
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47. https://doi.org/10.1093/nar/gkv007
Benjamini Y, Hochberg Y (1995) Controlling the false Discovery rate: a practical and powerful Approach to multiple testing. J Roy Stat Soc: Ser B (Methodol) 57(1):289–300. https://doi.org/10.1111/j.2517-6161.1995.tb02031.x
Blighe K (2021) EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. https://github.com/kevinblighe/EnhancedVolcano
Montgomery A, Tam F, Gursche C, Cheneval C, Besler K, Enns W, Manku S, Rey K, Hanson PJ, Rose-John S, McManus BM, Choy JC (2021) Overlapping and distinct biological effects of IL-6 classic and trans-signaling in vascular endothelial cells. Am J Physiol Cell Physiol 320(4):C554–C565. https://doi.org/10.1152/ajpcell.00323.2020
Zegeye MM, Lindkvist M, Falker K, Kumawat AK, Paramel G, Grenegard M, Sirsjo A, Ljungberg LU (2018) Activation of the JAK/STAT3 and PI3K/AKT pathways are crucial for IL-6 trans-signaling-mediated pro-inflammatory response in human vascular endothelial cells. Cell Commun Signal 16(1):55. https://doi.org/10.1186/s12964-018-0268-4
Rose-John S, Jenkins BJ, Garbers C, Moll JM, Scheller J (2023) Targeting IL-6 trans-signalling: past, present and future prospects. Nat Rev Immunol 1–16. https://doi.org/10.1038/s41577-023-00856-y
Ikonomidis I, Pavlidis G, Katsimbri P, Lambadiari V, Parissis J, Andreadou I, Tsoumani M, Boumpas D, Kouretas D, Iliodromitis E (2020) Tocilizumab improves oxidative stress and endothelial glycocalyx: a mechanism that may explain the effects of biological treatment on COVID-19. Food Chem Toxicol 145:111694. https://doi.org/10.1016/j.fct.2020.111694
Fels B, Acharya S, Vahldieck C, Graf T, Kading N, Rupp J, Kusche-Vihrog K (2022) Mineralocorticoid receptor-antagonism prevents COVID-19-dependent glycocalyx damage. Pflugers Arch 474(10):1069–1076. https://doi.org/10.1007/s00424-022-02726-3
Stahl K, Gronski PA, Kiyan Y, Seeliger B, Bertram A, Pape T, Welte T, Hoeper MM, Haller H, David S (2020) Injury to the endothelial glycocalyx in critically ill patients with COVID-19. Am J Respir Crit Care Med 202(8):1178–1181. https://doi.org/10.1164/rccm.202007-2676LE
Rovas A, Osiaevi I, Buscher K, Sackarnd J, Tepasse PR, Fobker M, Kuhn J, Braune S, Gobel U, Tholking G, Groschel A, Pavenstadt H, Vink H, Kumpers P (2021) Microvascular dysfunction in COVID-19: the MYSTIC study. Angiogenesis 24(1):145–157. https://doi.org/10.1007/s10456-020-09753-7
Mouffak S, Shubbar Q, Saleh E, El-Awady R (2021) Recent advances in management of COVID-19: a review. Biomed Pharmacother 143:112107. https://doi.org/10.1016/j.biopha.2021.112107
Aletaha D, Kerschbaumer A, Kastrati K, Dejaco C, Dougados M, McInnes IB, Sattar N, Stamm TA, Takeuchi T, Trauner M, van der Heijde D, Voshaar M, Winthrop KL, Ravelli A, Betteridge N, Burmester GR, Bijlsma JW, Bykerk V, Caporali R, Choy EH, Codreanu C, Combe B, Crow MK, de Wit M, Emery P, Fleischmann RM, Gabay C, Hetland ML, Hyrich KL, Iagnocco A, Isaacs JD, Kremer JM, Mariette X, Merkel PA, Mysler EF, Nash P, Nurmohamed MT, Pavelka K, Poor G, Rubbert-Roth A, Schulze-Koops H, Strangfeld A, Tanaka Y, Smolen JS (2023) Consensus statement on blocking interleukin-6 receptor and interleukin-6 in inflammatory conditions: an update. Ann Rheum Dis 82(6):773–787. https://doi.org/10.1136/ard-2022-222784
Group WHOREAC-TW, Shankar-Hari M, Vale CL, Godolphin PJ, Fisher D, Higgins JPT, Spiga F, Savovic J, Tierney J, Baron G, Benbenishty JS, Berry LR, Broman N, Cavalcanti AB, Colman R, De Buyser SL, Derde LPG, Domingo P, Omar SF, Fernandez-Cruz A, Feuth T, Garcia F, Garcia-Vicuna R, Gonzalez-Alvaro I, Gordon AC, Haynes R, Hermine O, Horby PW, Horick NK, Kumar K, Lambrecht BN, Landray MJ, Leal L, Lederer DJ, Lorenzi E, Mariette X, Merchante N, Misnan NA, Mohan SV, Nivens MC, Oksi J, Perez-Molina JA, Pizov R, Porcher R, Postma S, Rajasuriar R, Ramanan AV, Ravaud P, Reid PD, Rutgers A, Sancho-Lopez A, Seto TB, Sivapalasingam S, Soin AS, Staplin N, Stone JH, Strohbehn GW, Sunden-Cullberg J, Torre-Cisneros J, Tsai LW, van Hoogstraten H, van Meerten T, Veiga VC, Westerweel PE, Murthy S, Diaz JV, Marshall JC, Sterne JAC (2021) Association between Administration of IL-6 Antagonists and mortality among patients hospitalized for COVID-19: a Meta-analysis. JAMA 326(6):499–518. https://doi.org/10.1001/jama.2021.11330
Albuquerque AM, Eckert I, Tramujas L, Butler-Laporte G, McDonald EG, Brophy JM, Lee TC (2023) Effect of tocilizumab, sarilumab, and baricitinib on mortality among patients hospitalized for COVID-19 treated with corticosteroids: a systematic review and meta-analysis. Clin Microbiol Infect 29(1):13–21. https://doi.org/10.1016/j.cmi.2022.07.008
Godolphin PJ, Fisher DJ, Berry LR, Derde LPG, Diaz JV, Gordon AC, Lorenzi E, Marshall JC, Murthy S, Shankar-Hari M, Sterne JAC, Tierney JF, Vale CL (2022) Association between tocilizumab, sarilumab and all-cause mortality at 28 days in hospitalised patients with COVID-19: a network meta-analysis. PLoS ONE 17(7):e0270668. https://doi.org/10.1371/journal.pone.0270668
Yu SY, Koh DH, Choi M, Ryoo S, Huh K, Yeom JS, Yoon YK (2022) Clinical efficacy and safety of interleukin-6 receptor antagonists (tocilizumab and sarilumab) in patients with COVID-19: a systematic review and meta-analysis. Emerg Microbes Infect 11(1):1154–1165. https://doi.org/10.1080/22221751.2022.2059405
Pawar A, Desai RJ, Solomon DH, Santiago Ortiz AJ, Gale S, Bao M, Sarsour K, Schneeweiss S, Kim SC (2019) Risk of serious infections in tocilizumab versus other biologic drugs in patients with rheumatoid arthritis: a multidatabase cohort study. Ann Rheum Dis 78(4):456–464. https://doi.org/10.1136/annrheumdis-2018-214367
Murugesan H, Cs G, Nasreen HS, Santhanam S, M G, Ravi S, Es SS (2022) An evaluation of efficacy and safety of Tofacitinib, a JAK inhibitor in the management of hospitalized patients with mild to moderate COVID-19 - an open-label Randomized Controlled Study. J Assoc Physicians India 69(12):11–12
Guimaraes PO, Quirk D, Furtado RH, Maia LN, Saraiva JF, Antunes MO, Kalil Filho R, Junior VM, Soeiro AM, Tognon AP, Veiga VC, Martins PA, Moia DDF, Sampaio BS, Assis SRL, Soares RVP, Piano LPA, Castilho K, Momesso R, Monfardini F, Guimaraes HP, de Ponce D, Dulcine M, Pinheiro MRT, Gunay LM, Deuring JJ, Rizzo LV, Koncz T, Berwanger O, Investigators S-CT (2021) Tofacitinib in patients hospitalized with Covid-19 pneumonia. N Engl J Med 385(5):406–415. https://doi.org/10.1056/NEJMoa2101643
Sun J, Wang S, Ma X, Wei Q, Peng Y, Bai Y, Miao G, Meng C, Liu P (2023) Efficacy and safety of baricitinib for the treatment of hospitalized adults with COVID-19: a systematic review and meta-analysis. Eur J Med Res 28(1):536. https://doi.org/10.1186/s40001-023-01403-0
Hamilton FW, Thomas M, Arnold D, Palmer T, Moran E, Mentzer AJ, Maskell N, Baillie K, Summers C, Hingorani A, MacGowan A, Khandaker GM, Mitchell R, Davey Smith G, Ghazal P, Timpson NJ (2023) Therapeutic potential of IL6R blockade for the treatment of sepsis and sepsis-related death: a mendelian randomisation study. PLoS Med 20(1):e1004174. https://doi.org/10.1371/journal.pmed.1004174
Barkhausen T, Tschernig T, Rosenstiel P, van Griensven M, Vonberg RP, Dorsch M, Mueller-Heine A, Chalaris A, Scheller J, Rose-John S, Seegert D, Krettek C, Waetzig GH (2011) Selective blockade of interleukin-6 trans-signaling improves survival in a murine polymicrobial sepsis model. Crit Care Med 39(6):1407–1413. https://doi.org/10.1097/CCM.0b013e318211ff56
Greenhill CJ, Rose-John S, Lissilaa R, Ferlin W, Ernst M, Hertzog PJ, Mansell A, Jenkins BJ (2011) IL-6 trans-signaling modulates TLR4-dependent inflammatory responses via STAT3. J Immunol 186(2):1199–1208. https://doi.org/10.4049/jimmunol.1002971
Kang S, Onishi S, Ling Z, Inoue H, Zhang Y, Chang H, Zhao H, Wang T, Okuzaki D, Matsuura H, Takamatsu H, Oda J, Kishimoto T (2024) Gp130-HIF1alpha axis-induced vascular damage is prevented by the short-term inhibition of IL-6 receptor signaling. Proc Natl Acad Sci U S A 121(2):e2315898120. https://doi.org/10.1073/pnas.2315898120
Qiu X, Zhang L, Tong Y, Qu Y, Wang H, Mu D (2018) Interleukin-6 for early diagnosis of neonatal sepsis with premature rupture of the membranes: a meta-analysis. Med (Baltim) 97(47):e13146. https://doi.org/10.1097/MD.0000000000013146
Song J, Park DW, Moon S, Cho HJ, Park JH, Seok H, Choi WS (2019) Diagnostic and prognostic value of interleukin-6, pentraxin 3, and procalcitonin levels among sepsis and septic shock patients: a prospective controlled study according to the Sepsis-3 definitions. BMC Infect Dis 19(1):968. https://doi.org/10.1186/s12879-019-4618-7
Cong S, Ma T, Di X, Tian C, Zhao M, Wang K (2021) Diagnostic value of neutrophil CD64, procalcitonin, and interleukin-6 in sepsis: a meta-analysis. BMC Infect Dis 21(1):384. https://doi.org/10.1186/s12879-021-06064-0
Bode C, Weis S, Sauer A, Wendel-Garcia P, David S (2023) Targeting the host response in sepsis: current approaches and future evidence. Crit Care 27(1):478. https://doi.org/10.1186/s13054-023-04762-6
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the German Research Foundation (rotational position of KFO 342 – ZA428/18 − 1 to CD and AR; KU 2873/3 − 1 to PK) and the fund Innovative Medical Research of the University of Münster Medical School (I-RO221907 to AR).
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
CD designed and conducted the in vitro experiments, analyzed the data, prepared the figures and drafted the manuscript; AR and IO enrolled patients, performed sublingual video-microscopy, performed ELISA assays and contributed to the manuscript; KS contributed to statistical analysis and figure preparation; AL, HP and WL advised on the study design, discussed the findings and revised the manuscript; PK had the initial idea, supervised the study, recruited and coordinated participating centers, contributed to the analysis and revised the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was performed in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the General Medical Council Westfalen-Lippe and the WWU Münster, Germany (File Number: amendments of 2016-073-f-S). Patients were enrolled upon after obtaining written informed consent from them or their legal representatives.
Consent for publication
Consent for publication has been obtained.
Competing interests
All authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Drost, C.C., Rovas, A., Osiaevi, I. et al. Interleukin-6 drives endothelial glycocalyx damage in COVID-19 and bacterial sepsis. Angiogenesis (2024). https://doi.org/10.1007/s10456-024-09916-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10456-024-09916-w