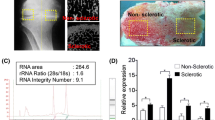
Abstract
Osteoarthritis (OA) is one of the most prevalent musculoskeletal diseases globally, leading to chronic disability and poor prognosis. One of the approaches for optimizing OA treatment is to find early effective diagnostic biomarkers. The contribution of microRNAs (miRNAs) in OA progression is now being increasingly recognized. This review provides a comprehensive summary on studies reporting the expression profiling of miRNAs in OA and associated signaling pathways. We performed a systematic search of the Embase, Web of Science, PubMed, and Cochrane library databases. This systematic review is reported according to the PRISMA checklist. Studies which identified miRNAs with aberrant expression compared to controls during OA progression were included, and a meta-analysis was performed. Results from the random effects model were provided as log10 odds ratios (logORs) and 95% confidence intervals. Sensitivity analysis was conducted to confirm the accuracy of the results. Subgroup analysis was conducted based on tissue source. The target genes of miRNAs identified in this study were extracted from the MiRWalk database, and these target genes were enriched in Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathways. A total of 191 studies reporting 162 miRNAs were included in our meta-analysis. Among them, 36 miRNAs distributed across 96 studies were expressed in the same direction in at least two studies (13 up-regulated and 23 down-regulated). Subgroup analysis of tissue source revealed that the highest number of studies was conducted using articular cartilage, where the most up-regulated miRNAs were miR-146a-5p (logOR 7.355; P < 0.001) and miR-34a-5p (logOR 6.955; P < 0.001), and the most down-regulated miRNAs were miR-127-5p (logOR 6.586; P < 0.001) and miR-140-5p (logOR 6.373; P < 0.001). Enrichment analysis of 752 downstream target genes of all identified miRNAs was performed, and the regulatory relationships among them were displayed. Mesenchymal stem cells and transforming growth factor-β were found to be the most important downstream effectors regulated by miRNA in OA. This study highlighted the importance of miRNA signaling in OA progression and identified a number of prominent miRNAs including miR-146a-5p, miR-34a-5p, miR-127-5p, and miR-140-5p which might be considered as potential biomarkers for OA.







Similar content being viewed by others
Data availability
The guarantor (BW) is willing to examine all requests for the full dataset after a period of two years from the date of this publication. The corresponding author should be contacted at BW. wangbin_pku@zju.edu.cn. The lead author (BW) affirms that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained. The datasets used and analyzed in this study are available upon reasonable request from the corresponding author.
Abbreviations
- OA:
-
Osteoarthritis
- MSCs:
-
Mesenchymal stem cells
- ncRNAs:
-
Non-coding RNAs
- circRNA:
-
Circular RNA
- lncRNA:
-
Long non-coding RNA
- miRNA:
-
MicroRNA
- mRNA:
-
Messenger RNA
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- QUADAS-2:
-
Quality Assessment of Diagnostic Accuracy Studies-2
- HRs:
-
Hazard ratios
- Cis:
-
Confidence intervals
- logOR:
-
Log-odds-ratio
- BP:
-
Biological processes
- CC:
-
Cellular component
- MF:
-
Molecular function
- MSCs:
-
Mesenchymal stem cells
- TGF:
-
Transforming growth factor
- VEGF:
-
Vascular endothelial growth factor
- BMP:
-
Bone morphogenetic protein
- MMP:
-
Matrix metalloproteinase
References
Safari R, Jackson J, Sheffield D. Digital self-management interventions for people with osteoarthritis: systematic review with meta-analysis. J Med Internet Res. 2020;22(7):e15365. https://doi.org/10.2196/15365.
Hunter DJ, Schofield D, Callander E. The individual and socioeconomic impact of osteoarthritis. Nature Rev Rheumatol. 2014;10(7):437–41. https://doi.org/10.1038/nrrheum.2014.44.
Fu M, Zhou H, Li Y, Jin H, Liu X. Global, regional, and national burdens of hip osteoarthritis from 1990 to 2019: estimates from the 2019 global burden of disease study. Arthritis Res Ther. 2022;24(1):8. https://doi.org/10.1186/s13075-021-02705-6.
Safiri S, Kolahi AA, Smith E, et al. Global, regional and national burden of osteoarthritis 1990–2017: a systematic analysis of the global burden of disease study 2017. Ann Rheum Dis. 2020;79(6):819–28. https://doi.org/10.1136/annrheumdis-2019-216515.
Kotlarz H, Gunnarsson CL, Fang H, Rizzo JA. Osteoarthritis and absenteeism costs: evidence from US national survey data. J Occup Environ Med. 2010;52(3):263–8. https://doi.org/10.1097/JOM.0b013e3181cf00aa.
Thysen S, Luyten FP, Lories RJ. Targets, models and challenges in osteoarthritis research. Dis Models Mech. 2015;8(1):17–30. https://doi.org/10.1242/dmm.016881.
Fernandes GS, Parekh SM, Moses J, et al. Prevalence of knee pain, radiographic osteoarthritis and arthroplasty in retired professional footballers compared with men in the general population: a cross-sectional study. Br J Sports Med. 2018;52(10):678–83. https://doi.org/10.1136/bjsports-2017-097503.
Berenbaum F, Wallace IJ, Lieberman DE, Felson DT. Modern-day environmental factors in the pathogenesis of osteoarthritis. Nat Rev Rheumatol. 2018;14(11):674–81. https://doi.org/10.1038/s41584-018-0073-x.
Kong H, Sun ML, Zhang XA, Wang XQ. Crosstalk among circRNA/lncRNA, miRNA, and mRNA in osteoarthritis. Front Cell Dev Biol. 2021;9:774370. https://doi.org/10.3389/fcell.2021.774370.
Li H, Yang HH, Sun ZG, Tang HB, Min JK. Whole-transcriptome sequencing of knee joint cartilage from osteoarthritis patients. Bone Joint Res. 2019;8(7):290–303. https://doi.org/10.1302/2046-3758.87.Bjr-2018-0297.R1.
Zhang W, Qi L, Chen R, et al. Circular RNAs in osteoarthritis: indispensable regulators and novel strategies in clinical implications. Arthritis Res Ther. 2021;23(1):23. https://doi.org/10.1186/s13075-021-02420-2.
Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(1):30. https://doi.org/10.1186/s12943-020-1135-7.
Chen C, Yin P, Hu S, Sun X, Li B. Circular RNA-9119 protects IL-1β-treated chondrocytes from apoptosis in an osteoarthritis cell model by intercepting the microRNA-26a/PTEN axis. Life Sci. 2020;256:117924. https://doi.org/10.1016/j.lfs.2020.117924.
Li L, Lv G, Wang B, Kuang L. The role of lncRNA XIST/miR-211 axis in modulating the proliferation and apoptosis of osteoarthritis chondrocytes through CXCR4 and MAPK signaling. Biochem Biophys Res Commun. 2018;503(4):2555–62. https://doi.org/10.1016/j.bbrc.2018.07.015.
Zhu Y, Li R, Wen LM. Long non-coding RNA XIST regulates chondrogenic differentiation of synovium-derived mesenchymal stem cells from temporomandibular joint via miR-27b-3p/ADAMTS-5 axis. Cytokine. 2021;137:155352. https://doi.org/10.1016/j.cyto.2020.155352.
Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5. https://doi.org/10.1038/nature02871.
Wang X, Ning Y, Zhou B, Yang L, Wang Y, Guo X. Integrated bioinformatics analysis of the osteoarthritis-associated microRNA expression signature. Mol Med Rep. 2018;17(1):1833–8. https://doi.org/10.3892/mmr.2017.8057.
Le LT, Swingler TE, Clark IM. Review: the role of microRNAs in osteoarthritis and chondrogenesis. Arthritis Rheum. 2013;65(8):1963–74. https://doi.org/10.1002/art.37990.
Le LT, Swingler TE, Crowe N, et al. The microRNA-29 family in cartilage homeostasis and osteoarthritis. J Mol Med (Berl). 2016;94(5):583–96. https://doi.org/10.1007/s00109-015-1374-z.
Ratneswaran A, Kapoor M. Osteoarthritis year in review: genetics, genomics, epigenetics. Osteoarthritis Cartil. 2021;29(2):151–60. https://doi.org/10.1016/j.joca.2020.11.003.
Li Z, Chen Z, Wang X, et al. Integrated analysis of miRNAs and gene expression profiles reveals potential biomarkers for osteoarthritis. Front Genet. 2022;13:814645. https://doi.org/10.3389/fgene.2022.814645.
Torres-Berrío A, Nouel D, Cuesta S, et al. MiR-218: a molecular switch and potential biomarker of susceptibility to stress. Mol Psychiatry. 2020;25(5):951–64. https://doi.org/10.1038/s41380-019-0421-5.
Xu J, Kang Y, Liao WM, Yu L. MiR-194 regulates chondrogenic differentiation of human adipose-derived stem cells by targeting Sox5. PLoS ONE. 2012;7(3):e31861. https://doi.org/10.1371/journal.pone.0031861.
Swingler TE, Wheeler G, Carmont V, et al. The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum. 2012;64(6):1909–19. https://doi.org/10.1002/art.34314.
Park SJ, Cheon EJ, Lee MH, Kim HA. MicroRNA-127–5p regulates matrix metalloproteinase 13 expression and interleukin-1beta-induced catabolic effects in human chondrocytes. Arthritis Rheum. 2013;65(12):3141–52. https://doi.org/10.1002/art.38188.
Liu JN, Lu S, Fu CM. MiR-146a expression profiles in osteoarthritis in different tissue sources: a meta-analysis of observational studies. J Orthop Surg Res. 2022;17(1):148. https://doi.org/10.1186/s13018-022-02989-7.
Jones TL, Esa MS, Li KHC, et al. Osteoporosis, fracture, osteoarthritis & sarcopenia: A systematic review of circulating microRNA association. Bone. 2021;152:116068. https://doi.org/10.1016/j.bone.2021.116068.
Shorter E, Avelar R, Zachariou M, et al. Identifying novel osteoarthritis-associated genes in human cartilage using a systematic meta-analysis and a multi-source integrated network. Int J Mol Sci. 2022. https://doi.org/10.3390/ijms23084395.
Swingler TE, Niu L, Smith P, et al. The function of microRNAs in cartilage and osteoarthritis. Clin Exp Rheumatol. 2019;37(5):40–7.
Gene Ontology C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015; 43(Database issue):D1049-56.https://doi.org/10.1093/nar/gku1179
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–62. https://doi.org/10.1093/nar/gkv1070.
Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–36. https://doi.org/10.7326/0003-4819-155-8-201110180-00009.
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58. https://doi.org/10.1002/sim.1186.
Newman MC. “What exactly are you inferring?” A closer look at hypothesis testing. Environ Toxicol Chem. 2008;27(5):1013–9. https://doi.org/10.1897/07-373.1.
Higgins JPTTJ, Chandler J, Cumpston M, Li T, Page MJ, Welch VA, editors. Cochrane handbook for systematic reviews of interventions version 6.3. Cochrane; 2022.
Sticht C, De La Torre C, Parveen A, Gretz N. miRWalk: an online resource for prediction of microRNA binding sites. PLoS ONE. 2018;13(10):e0206239. https://doi.org/10.1371/journal.pone.0206239.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics J Integr Biol. 2012;16(5):284–7. https://doi.org/10.1089/omi.2011.0118.
Tian F, Wang J, Zhang Z, Yang J. LncRNA SNHG7/miR-34a-5p/SYVN1 axis plays a vital role in proliferation, apoptosis and autophagy in osteoarthritis. Biol Res. 2020;53(1):9. https://doi.org/10.1186/s40659-020-00275-6.
Song P, Ye LF, Zhang C, Peng T, Zhou XH. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016;592(1):8–14. https://doi.org/10.1016/j.gene.2016.07.055.
Jia D, Niu Y, Li D, Liu Z. lncRNA C2dat1 promotes cell proliferation, migration, and invasion by targeting miR-34a-5p in osteosarcoma cells. Oncol Research. 2018;26(5):753–64. https://doi.org/10.3727/096504017x15024946480113.
Endisha H, Datta P, Sharma A, et al. MicroRNA-34a-5p promotes joint destruction during osteoarthritis. Arthritis Rheumatol. 2021;73(3):426–39. https://doi.org/10.1002/art.41552.
Dong C, Wang X, Li N, et al. microRNA-mediated GAS1 downregulation promotes the proliferation of synovial fibroblasts by PI3K-Akt signaling in osteoarthritis. Exp Ther Med. 2019;18(6):4273–86. https://doi.org/10.3892/etm.2019.8101.
Song AF, Kang L, Wang YF, Wang M. MiR-34a-5p inhibits fibroblast-like synoviocytes proliferation via XBP1. Eur Rev Med Pharmacol Sci. 2020;24(22):11675–82. https://doi.org/10.26355/eurrev_202011_23812.
Zhang H, Zheng W, Li D, Zheng J. miR-146a-5p promotes chondrocyte apoptosis and inhibits autophagy of osteoarthritis by targeting NUMB. Cartilage. 2021;13(2_suppl):1467s-s1477. https://doi.org/10.1177/19476035211023550.
Saito T, Fukai A, Mabuchi A, et al. Transcriptional regulation of endochondral ossification by HIF-2alpha during skeletal growth and osteoarthritis development. Nat Med. 2010;16(6):678–86. https://doi.org/10.1038/nm.2146.
Wang Y, Shen S, Li Z, Li W, Weng X. MIR-140–5p affects chondrocyte proliferation, apoptosis, and inflammation by targeting HMGB1 in osteoarthritis. Inflamm Res. 2020;69(1):63–73. https://doi.org/10.1007/s00011-019-01294-0.
Li C, Hu Q, Chen Z, et al. MicroRNA-140 suppresses human chondrocytes hypertrophy by targeting SMAD1 and controlling the bone morphogenetic protein pathway in osteoarthritis. Am J Med Sci. 2018;355(5):477–87. https://doi.org/10.1016/j.amjms.2018.01.004.
Karlsen TA, Jakobsen RB, Mikkelsen TS, Brinchmann JE. microRNA-140 targets RALA and regulates chondrogenic differentiation of human mesenchymal stem cells by translational enhancement of SOX9 and ACAN. Stem Cells Dev. 2014;23(3):290–304. https://doi.org/10.1089/scd.2013.0209.
Duan L, Liang Y, Xu X, Xiao Y, Wang D. Recent progress on the role of miR-140 in cartilage matrix remodelling and its implications for osteoarthritis treatment. Arthritis Res Ther. 2020;22(1):194. https://doi.org/10.1186/s13075-020-02290-0.
Liang Y, Duan L, Xiong J, et al. E2 regulates MMP-13 via targeting miR-140 in IL-1β-induced extracellular matrix degradation in human chondrocytes. Arthritis Res Ther. 2016;18(1):105. https://doi.org/10.1186/s13075-016-0997-y.
Li Z, Yuan B, Pei Z, et al. Circ_0136474 and MMP-13 suppressed cell proliferation by competitive binding to miR-127-5p in osteoarthritis. J Cell Mol Med. 2019;23(10):6554–64. https://doi.org/10.1111/jcmm.14400.
Tu M, Li Y, Zeng C, et al. MicroRNA-127-5p regulates osteopontin expression and osteopontin-mediated proliferation of human chondrocytes. Sci Rep. 2016;6:25032. https://doi.org/10.1038/srep25032.
Yan S, Wang M, Zhao J, et al. MicroRNA-34a affects chondrocyte apoptosis and proliferation by targeting the SIRT1/p53 signaling pathway during the pathogenesis of osteoarthritis. Int J Mol Med. 2016;38(1):201–9. https://doi.org/10.3892/ijmm.2016.2618.
Soyocak A, Kurt H, Ozgen M, Turgut Cosan D, Colak E, Gunes HV. miRNA-146a, miRNA-155 and JNK expression levels in peripheral blood mononuclear cells according to grade of knee osteoarthritis. Gene. 2017;627:207–11. https://doi.org/10.1016/j.gene.2017.06.027.
Shao JH, Ding ZR, Peng JH, et al. MiR-146a-5p promotes IL-1 beta-induced chondrocyte apoptosis through the TRAF6-mediated NF-kB pathway. Inflamm Res. 2020;69(6):619–30. https://doi.org/10.1007/s00011-020-01346-w.
Yuan C, Pan Z, Zhao K, et al. Classification of four distinct osteoarthritis subtypes with a knee joint tissue transcriptome atlas. Bone Res. 2020;8(1):38. https://doi.org/10.1038/s41413-020-00109-x.
Lv Z, Yang YX, Li J, et al. Molecular classification of knee osteoarthritis. Fron Cell Dev Biol. 2021;9:725568. https://doi.org/10.3389/fcell.2021.725568.
Skrzypa M, Szala D, Gablo N, et al. miRNA-146a-5p is upregulated in serum and cartilage samples of patients with osteoarthritis. Pol Przegl Chir. 2019;91(3):1–5. https://doi.org/10.5604/01.3001.0013.0135.
Zhang M, Wang M, Tan X, Li TF, Zhang YE, Chen D. Smad3 prevents beta-catenin degradation and facilitates beta-catenin nuclear translocation in chondrocytes. J Biol Chem. 2010;285(12):8703–10. https://doi.org/10.1074/jbc.M109.093526.
Li TF, Darowish M, Zuscik MJ, et al. Smad3-deficient chondrocytes have enhanced BMP signaling and accelerated differentiation. J Bone Miner Res Off J Am Soc Bone Miner Res. 2006;21(1):4–16. https://doi.org/10.1359/jbmr.050911.
Zhen G, Wen C, Jia X, et al. Inhibition of TGF-β signaling in mesenchymal stem cells of subchondral bone attenuates osteoarthritis. Nat Med. 2013;19(6):704–12. https://doi.org/10.1038/nm.3143.
Harrell CR, Markovic BS, Fellabaum C, Arsenijevic A, Volarevic V. Mesenchymal stem cell-based therapy of osteoarthritis: Current knowledge and future perspectives. Biomed Pharmacother. 2019;109:2318–26. https://doi.org/10.1016/j.biopha.2018.11.099.
Kroon LMG, Davidson ENB, Narcisi R, Farrell E, Kraan PMV, van Osch G. Activin and nodal are not suitable alternatives to TGFβ for chondrogenic differentiation of mesenchymal stem cells. Cartilage. 2017;8(4):432–8. https://doi.org/10.1177/1947603516667585.
Kou X, Xu X, Chen C, et al. The Fas/Fap-1/Cav-1 complex regulates IL-1RA secretion in mesenchymal stem cells to accelerate wound healing. Sci Transl Med. 2018. https://doi.org/10.1126/scitranslmed.aai8524.
Berthelot JM, Le Goff B, Maugars Y. Bone marrow mesenchymal stem cells in rheumatoid arthritis, spondyloarthritis, and ankylosing spondylitis: problems rather than solutions? Arthritis Res Ther. 2019;21(1):239. https://doi.org/10.1186/s13075-019-2014-8.
Funding
This study was supported by the National Natural Science Foundation of China (81802204) and by Zhejiang University School of Medicine, The First Affiliated Hospital’s Foundation (G2022010-18), Alibaba Cloud, Natural Science Foundation of Zhejiang Province (LTGY23H060007), and Zhejiang Medical and Health Science and Technology Project (2023RC010). The founders had no role in considering the study design or in the collection, analysis, interpretation of data, writing of the report, or decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
HCL, LY, and XKL contributed equally to this work. BW, HCL and LY conceived the idea for the review. BW, HCL, and LY designed, undertook the literature search, and coordinated the study. XKL, DJL, GSW, NNS, and JJL gave crucial intellectual input and provided critical revision for the initial protocol. XKL and DJL contributed to the implementation of the study. HCL, LY, and XKL acquired data, screened records, extracted data, and assessed risk of bias. HCL coded the statistical analysis, figures, and appendix in collaboration with LY and XKL. NNS, JJL, and BW analyzed and interpreted the data. HCL, LY, and BW wrote the first draft of the manuscript. All authors gave crucial feedback on the revised report and approved the final version of the manuscript. BW obtained funding. HCL, YL, XKL, and BW are the guarantors of this manuscript. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
Corresponding authors
Ethics declarations
Conflict of interest
All authors have completed the ICMJE uniform disclosure form at www.icmje.org/disclosure-of-interest/ and declare: All authors report no financial relationships with any organizations that might have an interest in the submitted work in the previous three years. All authors report no other relationships or activities that could appear to have influenced the submitted work.
Ethical approval
This is a systematic review and network meta-analysis and does not require ethical approval.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, H., Yan, L., Li, X. et al. MicroRNA expression in osteoarthritis: a meta-analysis. Clin Exp Med 23, 3737–3749 (2023). https://doi.org/10.1007/s10238-023-01063-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-023-01063-8