Abstract
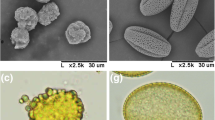
The thermo-sensitive genic male sterility (TGMS) system plays a key role in the production of two-line hybrids in rapeseed (Brassica napus). To uncover key cellular events and genetic regulation associated with TGMS, a combined study using cytological methods and RNA-sequencing analysis was conducted for the rapeseed TGMS line 373S. Cytological studies showed that microspore cytoplasm of 373S plants was condensed, the microspore nucleus was degraded at an early stage, the exine was irregular, and the tapetum developed abnormally, eventually leading to male sterility. RNA-sequencing analysis identified 430 differentially expressed genes (298 upregulated and 132 downregulated) between the fertile and sterile samples. Gene ontology analysis demonstrated that the most highly represented biological processes included sporopollenin biosynthetic process, pollen exine formation, and extracellular matrix assembly. Kyoto encyclopedia of genes and genomes analysis indicated that the enriched pathways included amino acid metabolism, carbohydrate metabolism, and lipid metabolism. Moreover, 26 transcript factors were identified, which may be associated with abnormal tapetum degeneration and exine formation. Subsequently, 19 key genes were selected, which are considered to regulate pollen development and even participate in pollen exine formation. Our results will provide important insight into the molecular mechanisms underlying TGMS in rapeseed.





Similar content being viewed by others
Data availability
The RNA-seq raw read data have been submitted in the Sequence Read Archive of the NCBI (accession number: PRJNA612938)
Code availability
Not applicable.
Abbreviations
- CHA:
-
chemical hybridization agent
- CMS:
-
cytoplasmic male sterility
- DAPI:
-
4′,6-diamidino-2-phenylindole
- DEGs:
-
differentially expressed genes
- EMS:
-
ecological male sterility
- FPKM:
-
fragments per kilobase of transcript per million mapped reads
- GMS:
-
genic male sterility
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- PMC:
-
pollen mother cell
- PTGMS:
-
photoperiod and temperature/thermo- sensitive genic male sterility
- P/TGMS:
-
photoperiod and/or temperature sensitive genic male sterility
- RGMS:
-
recessive genetic male sterility
- RIN:
-
RNA integrity number
- RNA-seq:
-
RNA-sequencing
- SI:
-
self-incompatibility
- TFs:
-
transcript factors
- TGMS:
-
thermo-sensitive genic male sterility
- ZS9:
-
Zhongshuang No. 9
References
Aarts MGM, Dirkse WG, Stiekema WJ, Pereira A (1993) Transposon tagging of a male sterility gene in Arabidopsis. Nature 363:715–717. https://doi.org/10.1038/363715a0
Aarts MGM, Hodge R, Kalantidis K, Florack D, Wilson ZA et al (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J 12:615–623. https://doi.org/10.1046/j.1365-313X.1997.00615.x
Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K (2003) A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol Biol 53:107–116. https://doi.org/10.1023/B:PLAN.0000009269.97773.70
Ariizumi T, Hatakeyama K, Hinata K, Inatsugi R, Nishida I, Sato S, Kato T, Tabata S, Toriyama K (2004) Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J 39:170–181. https://doi.org/10.1111/j.1365-313X.2004.02118.x
Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62:437–460. https://doi.org/10.1146/annurev-arplant-042809-112312
Chalhoub B, Denoeud F, Liu S et al (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345:950–953. https://doi.org/10.1126/science.1253435
Chen W, Yu XH, Zhang K, Shi J, De Oliveira S, Schreiber L, Shanklin J, Zhang D (2011) Male Sterile 2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol 157:842–853. https://doi.org/10.1104/pp.111.181693
Cock PJ, Fields CJ, Goto N, Heuer ML, Rice PM (2010) The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res 38:1767–1771. https://doi.org/10.1093/nar/gkp1137
De ASC, Kim SS, Koch S, Kienow L, Schneider K, McKim SM, Haughn GW, Kombrink E, Douglas CJ (2009) A novel fatty Acyl-coa synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21:507–525. https://doi.org/10.1105/tpc.108.062513
Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao L, Xu C, Li L, Xiao J, Zhang Q (2012) A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acda Sci USA 109:2654–2659. https://doi.org/10.1073/pnas.1121374109
Dobritsa AA, Shrestha J, Morant M, Pinot F, Matsuno M, Swanson R, Moller BL, Preuss D (2009) CYP704B1 is a long-chain fatty acid ω-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol 151:574–589. https://doi.org/10.1104/pp.109.144469
Dobritsa AA, Lei Z, Nishikawa S, Urbanczyk-Wochniak E, Huhman DV, Preuss D, Sumner LW (2010) LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol 153:937–955. https://doi.org/10.1104/pp.110.157446
Dong JG, Dong ZS, Liu XX, Liu CS, Li HB (2004) Cytological studies on anther development of ecological male sterile line 533S in Brassica napus L. J Northwest Sci-Tech Univ of Agri and For ( Nat Sci Ed ) 32:61–66. https://doi.org/10.1300/J064v24n01_09
Fan Y, Yang J, Mathioni SM, Yu J, Shen J, Yang X, Wang L, Zhang Q, Cai Z, Xu C, Li X, Xiao J, Meyers B, Zhang Q (2016) PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc Natl Acda Sci USA 113:15144–15149. https://doi.org/10.1073/pnas.1619159114
Frasch RM, Weigand C, Perez PT, Palmer RG, Sandhu D (2011) Molecular mapping of 2 environmentally sensitive male-sterile mutants in soybean. J Hered 102:11–16. https://doi.org/10.1093/jhered/esq100
Fu T (2019) Genetics and breeding of rapeseed hybrid. Hubei Science and technology press, Wuhan, Hubei, China
Gao H, Liu Z, Yuan W, Wu P, Zhou J (2010) Breeding of two-line hybrid rape variety Ganliangyou No. 3. Hubei Agric Sci 49:2370–2371. https://doi.org/10.1201/b10279-23
Grienenberger E, Kim SS, Lallemand B, Geoffroy P, Heintz D, Souza C, Heitz T, Douglas CJ, Legrand M (2010) Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22:4067–4083. https://doi.org/10.1105/tpc.110.080036
Higginson T, Li SF, Parish RW (2003) AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J 35:177–192. https://doi.org/10.1046/j.1365-313X.2003.01791.x
Ito T, Shinozaki K (2002) The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol 43:1285–1292. https://doi.org/10.1093/pcp/pcf154
Ito T, Nagata N, Yoshiba Y, Ohme-Takagi M, Ma H, Shinozaki K (2007) Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19:3549–3562. https://doi.org/10.1105/tpc.107.054536
Jia GX, Liu XD, Owen HA, Zhao DZ (2008) Signaling of cell fate determination by the TPD1 small protein and EMS1 receptor kinase. Proc Natl Acda Sci USA 105:2220–2225. https://doi.org/10.1073/pnas.0708795105
Jiang J, Zhang Z, Cao J (2013) Pollen wall development: the associated enzymes and metabolic pathways. Plant Biol 15:249–263. https://doi.org/10.1111/j.1438-8677.2012.00706.x
Jiang M, Zhu J, Wang W, Yang L (2022) Global transcriptome analysis reveals potential genes associated with genic male sterility of rapeseed (Brassica napus L.). Front Plant Sci 13:1004781. https://doi.org/10.3389/fpls.2022.1004781
Kim HU, Wu SSH, Ratnayake C, Huang AHC (2001) Brassica rapa has three genes that encode proteins associated with different neutral lipids in plastids of specific tissues. Plant Physiol 126:330–341. https://doi.org/10.1104/pp.126.1.330
Kim SS, Grienenberger E, Lallemand B, Colpitts CC, Kim SY, de Azevedo SC, Geoffroy P, Heintz D, Krahn D, Kaiser M, Kombrink E, Heitz T, Suh DY, Legrand M, Douglas CJ (2010) LAP6/POLYK ETIDE SYN THASE A and LAP5/POLYK ETIDE SYNTHASE B encode hydroxyalkyl α-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22:4045–4066. https://doi.org/10.1105/tpc.110.080028
Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, Wen TQ, Huang H, Luo D, Ma H, Zhang DB (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18:2999–3014. https://doi.org/10.1105/tpc.106.044107
Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, Franke R, Zhang P, Chen L, Gao Y, Liang W, Zhang D (2010) Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22:173–190. https://doi.org/10.4161/psb.5.9.12562
Liu ZW, Yuan WH, Xing LW, Wu P, Zhou XP, Zhou JG, Fu JH (2000) Breeding of two-line system hybrid Liangyou 586 in Brassica napus. Chin J Oil Crop Sci 22:5–7
Liu ZW, Wu P, Zhang QX, Zhou JG, Yuan WH, Zhou XP (2007) Breeding of high quality two-line hybrid rape variety Ganliangyou 2. Acta Agric Univ Jiangxiensis 19:10–11
Liu X, Huang J, Parameswaran S, Ito T, Seubert B, Auer M, Rymaszewski A, Jia G, Owen HA, Zhao D (2009) The SPOROCYTELESS/NOZZLE gene is involved in controlling stamen identity in Arabidopsis. Plant Physiol 151:1401–1411. https://doi.org/10.1104/pp.109.145896
Liu L, Fan XD (2013) Tapetum: regulation and role in sporopollenin biosynthesis in Arabidopsis. Plant Mol Biol 83:165–175. https://doi.org/10.1007/s11103-013-0085-5
Liu N, Wu P, Yuan W, Zhou J, Liu Z, Wang F (2013) Breeding of high-quality two-line hybrid rape variety Ganliangyou No. 5. Acta Agric Jiangxi 25:25–26
Liu XQ, Liu ZQ, Yu CY, Dong JG, Hu SW, Xu AX (2017) TGMS in rapeseed (Brassica napus) resulted in aberrant transcriptional regulation, asynchronous microsporocyte meiosis, defective tapetum, and fused sexine. Front Plant Sci 8:1268. https://doi.org/10.3389/fpls.2017.01268
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lu JY, Xiong SX, Yin W, Teng XD, Lou Y, Zhu J, Zhang C, Gu JN, Wilson ZA, Yang ZN (2020) MS1, a direct target of MS188, regulates the expression of key sporophytic pollen coat protein genes in Arabidopsis. J Exp Bot 71(16):4877–4889. https://doi.org/10.1093/jxb/eraa219
Ma H (2005) Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol 56:393–434. https://doi.org/10.1146/annurev.arplant.55.031903.141717
Ma XF, Wu Y, Zhang GF (2021) Formation pattern and regulatory mechanisms of pollen wall in Arabidopsis. J Plant Physiol 260:153388. https://doi.org/10.1016/j.jplph.2021.153388
Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449. https://doi.org/10.1093/bioinformatics/bti551
McCormick S (2004) Control of male gametophyte development. Plant Cell 16(Suppl):S142–S153. https://doi.org/10.1105/tpc.016659
McGettigan PA (2013) Transcriptomics in the RNA-seq era. Curr Opin Chem Biol 17:4–11. https://doi.org/10.1016/j.cbpa.2012.12.008
Morant M, Jorgensen K, Schaller H, Pinot F, Moller BL, Werck-Reichhart D, Bak S (2007) CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19:1473–1487. https://doi.org/10.1105/tpc.106.045948
Phan AH, Iacuone S, Li SF, Parish RW (2011) The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23:2209–2224. https://doi.org/10.1105/tpc.110.082651
Platt KA, Huang AHC, Thomson WW (1998) Ultrastructural study of lipid accumulation in tapetal cells of Brassica napus L. Cv. Westar during microsporogenesis. Int J Plant Sci 159:724–737. https://doi.org/10.2307/2475141
Qu C, Fu F, Liu M, Zhao H, Liu C, Li J, Tang Z, Xu X, Qiu X, Wang R, Lu K (2015) Comparative transcriptome analysis of recessive male sterility (RGMS) in sterile and fertile Brassica napus lines. PLoS One 10:e0144118. https://doi.org/10.1371/journal.pone.0144118
Quilichini TD, Douglas CJ, Samuels AL (2014) New views of tapetum ultrastructure and pollen exine development in Arabidopsis thaliana. Ann Bot 114:1189–1201. https://doi.org/10.1093/aob/mcu042
Rondanini DP, Gomez NV, Agosti MB, Miralles DJ (2012) Global trends of rapeseed grain yield stability and rapeseed-to-wheat yield ratio in the last four decades. Eur J Agron 37:56–65. https://doi.org/10.1016/j.eja.2011.10.005
Rowland O, Lee R, Franke R, Schreiber L, Kunst L (2007) The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Lett 581:3538–3544. https://doi.org/10.1016/j.febslet.2007.06.065
Ru ZG, Zhang LP, Hu TZ, Liu HY, Zhao CP (2015) Genetic analysis and chromosome mapping of a thermo-sensitive genic male sterile gene in wheat. Euphytica 201:321–327. https://doi.org/10.1007/s10681-014-1218-x
Rubinelli P, Hu Y, Ma H (1998) Identification, sequence analysis and expression studies of novel anther-specific genes of Arabidopsis thaliana. Plant Mol Biol 37:607–619. https://doi.org/10.1023/A:1005964431302
Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Yruong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11(29711):297. https://doi.org/10.1007/s004970050158
Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K (1999) Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acda Sci USA 96:11664–11669. https://doi.org/10.1073/pnas.96.20.11664
Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell 16(Suppl):S46–S60. https://doi.org/10.1016/j.laa.2009.09.016
Shi JX, Cui MH, Yang L, Kim YJ, Zhang DB (2015) Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci 20:741–753. https://doi.org/10.1016/j.tplants.2015.07.010
Sorensen AM, Krober S, Unte SU, Huijser P, Dekker K, Saedler H (2003) The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J 33:413–423. https://doi.org/10.1046/j.1365-313X.2003.01644.x
Sun X, Hu S, Yu C (2009) Cytological observation of anther development of an ecological male sterile line H50S in Brassica napus L. Acta Agric Bor-Occid Sin 18:153–158
Sun MX, Huang XY, Yang J, Guan YF, Yang ZN (2013) Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod 26:83–91. https://doi.org/10.1007/s00497-012-0208-1
Sun YY, Zhang DS, Wang ZZ, Guo Y, Sun XM, Li W, Zhi WL, Hu SW (2020) Cytological observation of anther structure and genetic investigation of a thermo-sensitive genic male sterile line 373S in Brassica napus L. BMC Plant Biol 20:8. https://doi.org/10.1186/s12870-019-2220-1
Sun YJ, Fu M, Wang L, Bai YX, Fang XL, Wang Q, He Y, Zeng HL (2022) OsSPLs regulate male fertility in response to different temperatures by flavonoid biosynthesis and tapetum PCD in PTGMS Rice. Int J Mol Sci 23:3744. https://doi.org/10.3390/ijms23073744
Tang JH, Fu ZY, Hu YM, Li JS, Sun LL, Ji HQ (2006) Genetic analyses and mapping of a new thermo-sensitive genic male sterile gene in maize. Theor Appl Genet 113:11. https://doi.org/10.1007/s00122-006-0262-x
Tang Z, Zhang L, Yang D, Zhao C, Zheng Y (2011) Cold stress contributes to aberrant cytokinesis during male meiosis I in a wheat thermosensitive genic male sterile line. Plant Cell Environ 34(3):389–405. https://doi.org/10.1111/j.1365-3040.2010.02250.x
Tang Z, Zhang L, Xu C, Yuan S, Zhang F, Zheng Y, Zhao C (2012) Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol 159(2):721–738. https://doi.org/10.1104/pp.112.196048
Tarazona S, García F, Ferrer A, Dopazo J, Conesa A (2011) Differential expression in RNA-seq: A matter of depth. Genome Res 21:2213–2223. https://doi.org/10.1101/gr.124321.111
Ting JTL, Wu SSH, Ratnayake C, Huang AHC (1998) Constituents of the tapetosomes and elaioplasts in Brassica campestris tapetum and their degradation and retention during microsporogenesis. Plant J 16:541–551. https://doi.org/10.1046/j.1365-313x.1998.00325.x
Truernit E, Stadler R, Baier K, Sauer N (1999) A male gametophyte-specific monosaccharide transporter in Arabidopsis. Plant J 17:191–201. https://doi.org/10.1046/j.1365-313X.1999.00372.x
Vizcay-Barrena G, Wilson ZA (2006) Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J Exp Bot 57:2709–2717. https://doi.org/10.1093/jxb/erl032
Wan XY and Li ZW (2019) Plant comparative transcriptomics reveals functional mechanisms and gene regulatory networks involved in anther development and male sterility. In: Blumenberg M (ed) Transcriptome analysis. https://doi.org/10.5772/intechopen.88318.77860
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63. https://doi.org/10.1038/nrg2484
Wang Y, Duan W, Bai J, Wang P, Yuan S, Zhao C, Zhang L (2019) Constitutive expression of a wheat microRNA, TaemiR167a, confers male sterility in transgenic Arabidopsis. Plant Growth Regul 88(3):227–239. https://doi.org/10.1007/s10725-019-00503-4
Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ (2001) The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J 28:27–39. https://doi.org/10.1046/j.1365-313x.2001.01125.x
Wu SSH, Moreau RA, Whitaker BD, Huang AHC (1999) Steryl esters in the elaioplasts of the tapetum in developing Brassica anthers and their recovery on the pollen surface. Lipids 34:517–523. https://doi.org/10.1007/s11745-999-0393-5
Wu LY, Jin XH, Zhang BL, Chen SJ, Xu R, Duan PG, Zou DN, Huang SJ, Zhou TB, An CC, Luo YH, Li YH (2022) A natural allele of OsMS1 responds to temperature changes and confers thermosensitive genic male sterility. Nat Commun 13(1):2055. https://doi.org/10.1038/s41467-022-29648-z
Xiao Q, Wang HD, Chen H, Wen J, Dai C, Ma CZ, Tu JX, Shen JX, Fu TD, Yi B (2021) Molecular analysis uncovers the mechanism of fertility restoration in temperature-sensitive Polima cytoplasmic male-sterile Brassica napus. Int J Mol Sci 22:12450. https://doi.org/10.3390/ijms222212450
Xing M, Guan C, Guan M (2022) Comparative cytological and transcriptome analyses of anther development in Nsa cytoplasmic male sterile (1258A) and maintainer lines in Brassica napus produced by distant hybridization. Int J Mol Sci 23:2004. https://doi.org/10.3390/ijms23042004
Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, Liang W, Zhang D, Wilson ZA (2010) The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22:91–107. https://doi.org/10.1105/tpc.109.071803
Xu J, Ding Z, Vizcay-Barrena G, Shi J, Liang W, Yuan Z, Werck-Reichhart D, Schreiber L, Wilson ZA, Zhang D (2014a) ABORTED MICROSPORES acts as a master regulator of pollen wall formation in Arabidopsis. Plant Cell 26:1544–1556. https://doi.org/10.1105/tpc.114.122986
Xu XF, Hu YM, Yu CY, Ge J, Guo YF, Dong JG, Hu SW (2014b) Physiological characterization and genetic analysis of reverse thermo-sensitive genic male-sterile line Huiyou 50S in Brassica napus. Acta Agric Boreal Sin 29:147–152
Xu D, Shi J, Rautengarten C, Yang L, Qian X, Uzair M, Zhu L, Luo Q, An G, Wassmann F, Schreiber L, Heazlewood JL, Scheller HV, Hu J, Zhang D, Liang W (2017) Defective Pollen wall 2 (DPW2) encodes an acyl transferase required for rice pollen development. Plant Physiol 173:240–255. https://doi.org/10.1104/pp.16.00095
Yan XH, Zeng XH, Wang SS, Li KQ, Yuan R, Gao HF, Luo JL, Liu F, Wu YH, Li YJ, Zhu L, Wu G (2016) Aberrant meiotic prophase I leads to genic male sterility in the novel TE5A mutant of Brassica napus. Sci Rep 6:33955. https://doi.org/10.1038/srep33955
Yang WC, Ye D, Xu J, Sundaresan V (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13:2108–2117. https://doi.org/10.1101/gad.13.16.2108
Yang SL, Xie LF, Mao HZ, Pauh CS, Yang WC, Jiang L, Sundaresan V, Ye D (2003) TAPETUM DETERMINANT 1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15:2792–2804. https://doi.org/10.1105/tpc.016618
Yang SL, Jiang L, Puah CS, Xie LF, Zhang XQ, Chen LQ, Yang WC, Ye D (2005) Overexpression of TAPETUM DETERMINANT1 alters the cell fates in the Arabidopsis carpel and tapetum via genetic interaction with EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS. Plant Physiol 139:186–191. https://doi.org/10.1104/pp.105.063529
Yang C, Vizcay-Barrena G, Conner K, Wilson ZA (2007) MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19:3530–3548. https://doi.org/10.1105/tpc.107.054981
Yang X, Wu D, Shi J, He Y, Pinot F, Grausem B, Yin C, Zhu L, Chen M, Luo Z, Liang W, Zhang D (2014) Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J Integr Plant Biol 56:979–994. https://doi.org/10.1111/jipb.12212
Yang XT, Ye JL, Zhang LL, Song XY (2020) Blocked synthesis of sporopollenin and jasmonic acid leads to pollen wall defects and anther indehiscence in genic male sterile wheat line 4110S at high temperatures. Funct Integr Genomic 20:383–396. https://doi.org/10.1007/s10142-019-00722-y
Yu CY, Li W, Chang JJ, Hu SW (2007) Development of a thermo-sensitive male-sterile line 373S in Brassica napus L. Chin Agric Sci Bull 23:245–248. https://doi.org/10.3969/j.issn.1000-6850.2007.07.057
Yu CY, Guo YF, Ge J, Hu YM, Dong JG, Dong ZS (2015) Characterization of a new temperature-sensitive male sterile line SP2S in rapeseed (Brassica napus L.). Euphytica 206:473–485. https://doi.org/10.1007/s10681-015-1514-0
Yuan GL, Wang YK, Yuan SH, Wang P, Duan WJ, Bai JF, Sun H, Wang N, Zhang FT, Zhang LP, Zhao CP (2018) Functional Analysis of wheat TaPaO1 gene conferring pollen sterility under low temperature. J Plant Biol 61(1):25–32. https://doi.org/10.1007/s12374-017-0269-7
Zhao DZ, Wang GF, Speal B, Ma H (2002) The EXCESS MICROSPOROCYTES1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev 16:2021–2031. https://doi.org/10.1101/gad.997902
Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H (2006) Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133:3085–3095. https://doi.org/10.1242/dev.02463
Zhang ZB, Zhu J, Gao JF, Wang C, Li H, Li H, Zhang HQ, Zhang S, Wang DM, Wang QX, Huang H, Xia HJ, Yang ZN (2007) Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J 52:528–538. https://doi.org/10.1111/j.1365-313x.2007.03254.x
Zhang C, Xu T, Ren MY, Zhu J, Shi QS, Zhang YF, Qi YW, Huang MJ, Song L, Xu P, Yang ZN (2020a) Slow development restores the fertility of photoperiod-sensitive male-sterile plant lines. Plant Physiol 184:923–932. https://doi.org/10.1104/pp.20.00951
Zhang M, Liu J, Ma Q, Qin Y, Wang HT, Chen PY, Ma L, Fu XK, Zhu LF, Wei HL, Yu SX (2020b) Deficiencies in the formation and regulation of anther cuticle and tryphine contribute to male sterility in cotton PGMS line. BMC Genomics 21:825. https://doi.org/10.21203/rs.3.rs-18152/v1
Zhou H, Zhou M, Yang Y et al (2014) RNase ZS1 processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice. Nat Commun 5:4884. https://doi.org/10.1038/ncomms5884
Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN (2008) Defective in Tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J 55:266–277. https://doi.org/10.1111/j.1365-313X.2008.03500.x
Zhu J, Lou Y, Xu X, Yang ZN (2011) A genetic pathway for tapetum development and function in Arabidopsis. J Integr Plant Biol 53:892–900. https://doi.org/10.1111/j.1744-7909.2011.01078.x
Zhu J, Lou Y, Shi QS, Zhang S, Zhou WT, Yang J, Zhang C, Yao XZ, Xu T, Liu JL (2020) Slowing development restores the fertility of thermos-sensitive male-sterile plant lines. Nature Plants 6:360–367. https://doi.org/10.1038/s41477-020-0622-6
Acknowledgements
The authors thank Prof. Huixian Zhao, College of Life Sciences, Northwest A&F University, for her suggestions in the course of preparing the manuscript. We are also grateful to our anonymous reviewers for their thoughtful and detailed reviews.
Funding
This research was funded by the National Natural Science Foundation of China (3217150949), the Key Research and Development Project in Shaanxi Province of China (2018ZDXM-NY-008; 2022NY-153), Modern Crop Seed Industry Project of Shaanxi Province (20171010000004), and the Key Research and Development Projects of Yangling Seed Industry Innovation Center (YLzy-yc2021-01).
Author information
Authors and Affiliations
Contributions
SH conceived and designed research. YS and DZ performed the experiments. YS and SH analyzed the data. YS and SH wrote the manuscript. HD, ZW, JW, HL and YG discussed and edited the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
The list of differentially expressed genes in pairwise comparisons between the fertile and sterile parent, as well as between fertile and sterile individuals in BC1 in two stages (bud and anther) (XLSX 1074 kb)
Additional file 2:
The 430 differentially expressed genes and their expression pattern in different stages (XLSX 123 kb)
Additional file 3:
Annotation of the 430 differentially expressed genes (XLSX 42 kb)
Additional file 4:
Gene ontology (GO) terms of each cluster in the heatmap (XLSX 261 kb)
Additional file 5:
Results of gene ontology term enrichment of the 430 differentially expressed genes (XLSX 18 kb)
Additional file 6:
The known and predicted interactions among the differentially expressed genes by String (XLSX 10 kb)
Additional file 7:
Cis-regulatory element analysis of the 430 differentially expressed genes (XLSX 1092 kb)
Additional file 8:
RNA-seq of DEGs in 373S under restrictive condition (high temperature) and permissive condition (low temperature) (XLSX 50 kb)
Additional file 9:
Fig. S1. Analysis of the relationship among all 15 samples according to RNA-seq based gene expression values. (A) The heatmap of sample correlation. The bar in right represents the scale of the relationship among samples, and the value in each pane represents the correlation coefficient between two samples. (B) Principal components analysis. All samples were clustered into two separate groups corresponding to developmental stages, buds and anthers shown by cyan and hermosa pink colors, respectively. The numbers in parentheses after PC (principal component) represent the proportion of variance explained by that PC. (C) The variance explained by the first four PCs. (PNG 298 kb)

Additional file 10:
Fig. S2. The pair-wise comparisons between fertile and sterile samples. The criteria for screening DEGs were probability≥0.8 and log2(fold change)≥1. 5A219F-A, anthers from fertile plants in BC1 (5A214// 5A214/5C95); 5A219F-B, buds from fertile plants in BC1; 5A219S-A, anthers from sterile plants in BC1; 5A219S-B, buds from sterile plants in BC1; 5C95-A, anthers from the fertile parent; 5C95-B, buds from the fertile parent; 5A214-A, anthers from the sterile parent; 5A214-B, buds from the sterile parent. (PNG 91 kb)

Additional file 11:
Fig. S3. Venn diagrams showing distribution of 430 differentially expressed genes between male fertile and sterile samples (PNG 50 kb)

Additional file 12:
Fig. S4. Classification of gene ontology annotations. The x-axis indicates categories and sub-categories, and the y-axis the number of genes. (PNG 395 kb)

Additional file 13:
Fig. S5. A network of differentially regulated gene ontology (GO) terms in biological processes. The scheme was drawn by using the BiNGO plugin of Cytoscape software. GO terms are presented as nodes and associations as edges, node size represents gene number, the bigger of the node size representing the more gene. The significant level of nodes is shown by different yellow colors as indicated in the scale bar. The lines with arrows mean the regulating relation. (PNG 491 kb)

Additional file 14:
Fig. S6. Gene ontology functional enrichment of differential expressed genes in biological process. Coloring indicates p-value (high: yellow, low: red). The lower p-value indicates the more significant enrichment. The lines with arrows mean the regulating relation. (PNG 276 kb)

Additional file 15:
Fig. S7. A network of protein-protein interactions based on analysis of the differentially expressed genes. The scheme was drawn using the STRING software. Genes are shown as nodes and interactions as edges (PNG 2745 kb)

Additional file 16:
Fig. S8. The most enriched pathways of the differentially expressed genes (DEGs) between sterile and fertile samples. (A) The most enriched metabolic activity between the sterile and fertile samples. (B) Pathway enrichment of the DEGs. The y-axis shows significantly enriched kyoto encyclopedia of genes and genomes pathways, and the x-axis shows the rich factor. Rich factor stands for the ratio of the number of DEGs belonging to a pathway to the number of all the annotated genes located in the pathway. The higher rich factor represents the higher level of enrichment. The size of the dot indicates the number of DEGs in the pathway, and the color of the dot reflects the different p-value range. (PNG 412 kb)

Additional file 17:
Fig. S9. Cluster analysis of 19 differentially expressed genes associated with pollen development. The color key represents log10 (FPKM+1). Color intensities (from green to red shading) increase with elevated expression level, as indicated at upper left. (PNG 207 kb)
Additional file 18:
Table S1. Description of the samples in the present study (PDF 39 kb)
Additional file 19:
Table S2. Target genes and their primer sequences for qRT-PCR (PDF 22 kb)
Additional file 20:
Table S3. Tetrad size and callose thickness in fertile control and 373S male sterile plants (PDF 104 kb)
Additional file 21:
Table S4. The statistics of RNA-sequencing data in individual samples (PDF 48 kb)
Additional file 22
Table S5 Verification of 430 DEGs identified between sterile samples and fertile samples by the RNA-seq experiment of 373S line under restrictive condition (high temperature) and permissive condition (low temperature) (PDF 106 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, Y., Zhang, D., Dong, H. et al. Comparative transcriptome analysis provides insight into the important pathways and key genes related to the pollen abortion in the thermo-sensitive genic male sterile line 373S in Brassica napus L.. Funct Integr Genomics 23, 26 (2023). https://doi.org/10.1007/s10142-022-00943-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-022-00943-8