Abstract
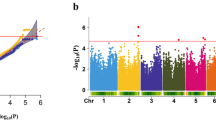
Starch is an important primary metabolite in plants, which can provide bioenergy for fuel ethanol production. There are many studies focusing on starch metabolism in Arabidopsis, maize, and rice, but few reports have been made on the starch content of tobacco leaves. Hence, to identify the marker-trait associations and isolate the candidate genes related to starch content of tobacco leaf, the genome-wide association study (GWAS) was performed using a multiparent advanced generation intercross (MAGIC) population consisting of 276 accessions genotyped by a 430 K SNP array. In this study, we detected the leaf starch content of tobacco plants cultivated in two places (Zhucheng and Chenzhou), which showed a wide variation of starch content in the population. A total of 28 and 45 significant single-nucleotide polymorphism (SNP) loci associated with leaf starch content were identified by single-locus and multi-locus GWAS models, respectively, and the phenotypic variance explained by these loci varied from 1.80 to − 14.73%. Furthermore, among these quantitative trait loci (QTLs), one SNP, AX-106011713 located on chromosome 19, was detected repeatedly in multiple models and two environments, which was selected for linkage disequilibrium (LD) analysis to obtain the target candidate region. Through gene annotation, haplotype, and gene expression analysis, two candidate genes encoding E3 ubiquitin-protein ligase (Ntab0823160) and fructose-bisphosphate aldolase (Ntab0375050) were obtained. Results showed that the variety carrying the beneficial alleles of the two candidate genes had higher gene expression level and leaf starch content, suggesting the potential role of candidate genes in enhancing the level of tobacco leaf starch content. Furthermore, silencing of Ntab0823160 in tobacco leaves reduced the content of total starch to 39.41–69.75% of that in the wide type plants. Taken together, our results provide useful resources for further investigation of the starch metabolic pathway and are also beneficial for the creation of eco-friendly cultivars with increased accumulation of leaf starch content.








Similar content being viewed by others
Abbreviations
- GWAS:
-
Genome-wide association study
- LD:
-
Linkage disequilibrium
- QTL:
-
Quantitative trait locus
- SNP:
-
Single-nucleotide polymorphism
- MAGIC:
-
Multiparent advanced generation intercross
References
Andersson M, Turesson H, Arrivault S et al (2018) Inhibition of plastid PPase and NTT leads to major changes in starch and tuber formation in potato. J Exp Bot 69:1913–1924. https://doi.org/10.1093/jxb/ery051
Aoyama S, Terada S, Sanagi M et al (2017) Membrane-localized ubiquitin ligase ATL15 functions in sugar-responsive growth regulation in Arabidopsis. Biochem Biophys Res Commun 491:33–39. https://doi.org/10.1016/j.bbrc.2017.07.028
Andrianov V, Borisjuk N, Pogrebnyak N et al (2010) Tobacco as a production platform for biofuel: overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol J 8:277–287. https://doi.org/10.1111/j.1467-7652.2009.00458.x
Asatsuma S, Sawada C, Itoh K et al (2005) Involvement of α-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol 46:858–869. https://doi.org/10.1093/pcp/pci091
Baker RF, Braun DM (2007) tie-dyed1 functions non-cell autonomously to control carbohydrate accumulation in maize leaves. Plant Physiol 144:867–878. https://doi.org/10.1104/pp.107.098814
Baunsgaard L, Lütken H, Mikkelsen R et al (2005) A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated α-glucans and is involved in starch degradation in Arabidopsis. Plant J 41:595–605. https://doi.org/10.1111/j.1365-313X.2004.02322.x
Bradbury PJ, Zhang Z, Kroon DE et al (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Boehlein SK, Shaw JR, Boehlein TJ et al (2018) Fundamental differences in starch synthesis in the maize leaf, embryo, ovary and endosperm. Plant J 96:595–606. https://doi.org/10.1111/tpj.14053
Burk DH, Liu B, Zhong R et al (2001) A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13:807–827
Cai Y, Li S, Jiao G et al (2018) OsPK2 encodes a plastidic pyruvate kinase involved in rice endosperm starch synthesis, compound granule formation and grain filling. Plant Biotechnol J 16:1878–1891. https://doi.org/10.1111/pbi.12923
Carrari F, Coll-Garcia D, Schauer N et al (2005) Deficiency of a plastidial adenylate kinase in arabidopsis results in elevated photosynthetic amino acid biosynthesis and enhanced growth. Plant Physiol 137:70–82. https://doi.org/10.1104/pp.104.056143
Chen Y, Gao Y, Li T et al (2021) Overexpression of NtAGPase small subunit gene increases leaf starch content and tobacco biomass. Shengwu Gongcheng Xuebao Chinese J Biotechnol 37:2845–2855. https://doi.org/10.13345/j.cjb.210149
Chen S, Hajirezaei M, Börnke F (2005) Differential expression of sucrose-phosphate synthase isoenzymes in tobacco reflects their functional specialization during dark-governed starch mobilization in source leaves. Plant Physiol 139:1163–1174. https://doi.org/10.1104/pp.105.069468
Corbesier L, Lejeune P, Bernier G (1998) The role of carbohydrates in the induction of flowering thaliana: comparison between the wild type and a star. Planta 206:131–137
Dong X, Gao Y, Chen W et al (2015) Spatiotemporal distribution of phenolamides and the genetics of natural variation of hydroxycinnamoyl spermidine in rice. Mol Plant 8:111–121. https://doi.org/10.1016/j.molp.2014.11.003
Emes MJ, Bowsher CG, Hedley C et al (2003) Starch synthesis and carbon partitioning in developing endosperm. J Exp Bot 54:569–575. https://doi.org/10.1093/jxb/erg089
Fulton DC, Stettler M, Mettler T et al (2008) β-Amylase4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. Plant Cell 20:1040–1058. https://doi.org/10.1105/tpc.107.056507
George GM, van der Merwe MJ, Nunes-Nesi A et al (2010) Virus-induced gene silencing of plastidial soluble inorganic pyrophosphatase impairs essential leaf anabolic pathways and reduces drought stress tolerance in Nicotiana benthamiana. Plant Physiol 154:55–66. https://doi.org/10.1104/pp.110.157776
Haake V, Zrenner R, Sonnewald U, Stitt M (1998) A moderate decrease of plastid aldolase activity inhibits photosynthesis, alters the levels of sugars and starch, and inhibits growth of potato plants. Plant J 14:147–157
Hanson KR, Mchale NA (1988) A starchless mutant of Nicotiana sylvestris containing a modified plastid phosphoglucomutase. Plant Physiol 88:838–844
Hirose T, Aoki N, Harada Y et al (2013) Disruption of a rice gene for α-glucan water dikinase, OsGWD1, leads to hyperaccumulation of starch in leaves but exhibits limited effects on growth. Front Plant Sci 4:1–9. https://doi.org/10.3389/fpls.2013.00147
Hu S, Wang M, Zhang X, et al (2021) Genetic basis of kernel starch content decoded in a maize multi-parent population. Plant Biotechnol J 19:2192–2205. https://doi.org/10.1111/pbi.13645
Huang BE, Verbyla KL, Verbyla AP et al (2015) MAGIC populations in crops: current status and future prospects. Theor Appl Genet 128:999–1017. https://doi.org/10.1007/s00122-015-2506-0
Huang LF, Liu YK, Su SC et al (2020) Genetic engineering of transitory starch accumulation by knockdown of OsSEX4 in rice plants for enhanced bioethanol production. Biotechnol Bioeng 117:933–944. https://doi.org/10.1002/bit.27262
James MG, Denyer K, Myers AM (2003) Starch synthesis in the cereal endosperm. Curr Opin Plant Biol 6:215–222. https://doi.org/10.1016/S1369-5266(03)00042-6
Khan SU, Yangmiao J, Liu S, et al (2019) Genome-wide association studies in the genetic dissection of ovule number, seed number, and seed weight in Brassica napus L. Ind Crops Prod 142:111877. https://doi.org/10.1016/j.indcrop.2019.111877
Kötting O, Santelia D, Edner C et al (2009) STARCH-EXCESS4 is a laforin-like phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell 21:334–346. https://doi.org/10.1105/tpc.108.064360
Kover PX, Valdar W, Trakalo J, et al (2009) A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet 5:e1000551. https://doi.org/10.1371/journal.pgen.1000551
Li N, Lin B, Wang H et al (2019) Natural variation in Zm FBL41 confers banded leaf and sheath blight resistance in maize. Nat Genet 51:1540–1548. https://doi.org/10.1038/s41588-019-0503-y
Lin MT, Occhialini A, Andralojc PJ et al (2014) A faster Rubisco with potential to increase photosynthesis in crops. Nature 513:547–550. https://doi.org/10.1038/nature13776
Lin TP, Caspar T, Somerville C, Preiss J (1988) Isolation and characterization of a starchless mutant of Arabidopsis thaliana (L.) Heynh lacking ADP glucose pyrophosphorylase activity. Plant Physiol 86:1131–1135
Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30:415–429. https://doi.org/10.1046/j.1365-313X.2002.01297.x
Luo J (2015) Metabolite-based genome-wide association studies in plants. Curr Opin Plant Biol 24:31–38. https://doi.org/10.1016/j.pbi.2015.01.006
MacNeill GJ, Mehrpouyan S, Minow MAA et al (2017) Starch as a source, starch as a sink: the bifunctional role of starch in carbon allocation. J Exp Bot 68:4433–4453. https://doi.org/10.1093/jxb/erx291
Mérida A, Fettke J (2021) Starch granule initiation in Arabidopsis thaliana chloroplasts. Plant J 107:688–697. https://doi.org/10.1111/tpj.15359
Niittylä T, Messerli G, Trevisan M et al (2004) A previously unknown maltose transporter essential for starch degradation in leaves. New Ser 303:87–89
Paul MJ, Foyer CH (2001) Sink regulation of photosynthesis. J Exp Bot 52:1383–1400
Peng M, Gao Y, Chen W et al (2016) Evolutionarily distinct BAHD N-acyltransferases are responsible for natural variation of aromatic amine conjugates in rice. Plant Cell 28:1533–1550. https://doi.org/10.1105/tpc.16.00265
Reid WV, Ali MK, Field CB (2020) The future of bioenergy. Glob Chang Biol 26:274–286. https://doi.org/10.1111/gcb.14883
Ritte G, Lloyd JR, Eckermann N et al (2002) The starch-related R1 protein is an-glucan, water dikinase. Proc Natl Acad Sci 99:7166–7171
Sallam A, Martsch R (2015) Association mapping for frost tolerance using multi-parent advanced generation inter-cross (MAGIC) population in faba bean (Vicia faba L.). Genetica 143:501–514. https://doi.org/10.1007/s10709-015-9848-z
Sanz-Barrio R, Corral-Martinez P, Ancin M et al (2013) Overexpression of plastidial thioredoxin f leads to enhanced starch accumulation in tobacco leaves. Plant Biotechnol J 11:618–627. https://doi.org/10.1111/pbi.12052
Sarah S, Moffatt BA (2004) Applying high throughput techniques in the study of adenosine kinase in plant metabolism and development. Front Biosci 9:1771–1781
Schulze W, Stitt M, Schulze E-D et al (1991) A quantification of the significance of assimilatory starch for growth of Arabidopsis thaliana L. Heynh. Plant Physiol 95:890–895
Schönhals EM, Ding J, Ritter E et al (2017) Physical mapping of QTL for tuber yield, starch content and starch yield in tetraploid potato (Solanum tuberosum L.) by means of genome wide genotyping by sequencing and the 8.3 K SolCAP SNP array. BMC Genomics 18:1–20. https://doi.org/10.1186/s12864-017-3979-9
Si L, Chen J, Huang X et al (2016) OsSPL13 controls grain size in cultivated rice. Nat Genet 48:447–456. https://doi.org/10.1038/ng.3518
Slewinski TL, Meeley R, Braun DM (2009) Sucrose transporter1 functions in phloem loading in maize leaves. J Exp Bot 60:881–892. https://doi.org/10.1093/jxb/ern335
Somerville C (2007) Biofuels. Curr Biol 17:115–119. https://doi.org/10.1016/j.cub.2007.01.010
Stitt M, Zeeman SC (2012) Starch turnover: pathways, regulation and role in growth. Curr Opin Plant Biol 15:282–292. https://doi.org/10.1016/j.pbi.2012.03.016
Sun C, Palmqvist S, Olsson H et al (2003) A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15:2076–2092. https://doi.org/10.1105/tpc.014597
Sun Y, Jiang Z, Liu H et al (2020) Genome-wide association analysis of tobacco flowering time. Chinese Tob Sci 41:1–6
Tamoi M, Nagaoka M, Miyagawa Y, Shigeoka S (2006) Contribution of fructose-1,6-bisphosphatase and sedoheptulose-1,7-bisphosphatase to the photosynthetic rate and carbon flow in the Calvin cycle in transgenic plants. Plant Cell Physiol 47:380–390. https://doi.org/10.1093/pcp/pcj004
Tieman D, Zhu G, Resende MFR et al (2017) Plant Science A chemical genetic roadmap to improved tomato flavor Downloaded from. Science (80- ) 355:391–394
Tsai HL, Lue WL, Lu KJ et al (2009) Starch synthesis in Arabidopsis is achieved by spatial cotranscription of core starch metabolism genes. Plant Physiol 151:1582–1595. https://doi.org/10.1104/pp.109.144196
Uematsu K, Suzuki N, Iwamae T et al (2012) Increased fructose 1,6-bisphosphate aldolase in plastids enhances growth and photosynthesis of tobacco plants. J Exp Bot 63:3001–3009. https://doi.org/10.1093/jxb/ers004
Wang RH, Wang TY, Li Y (2007) Linkage disequilibrium in plant genomes. Hereditas 29:1317–1323. https://doi.org/10.1360/yc-007-1317
Wen W, Li D, Li X et al (2014) Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun 5:1–10. https://doi.org/10.1038/ncomms4438
Xiao Q, Wang Y, Du J et al (2017) ZmMYB14 is an important transcription factor involved in the regulation of the activity of the ZmBT1 promoter in starch biosynthesis in maize. FEBS J 284:3079–3099. https://doi.org/10.1111/febs.14179
Yano K, Yamamoto E, Aya K et al (2016) Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat Genet 48:927–934. https://doi.org/10.1038/ng.3596
Yu TS, Lue W-L, Wang S-M, Chen J (2000) Mutation of Arabidopsis plastid phosphoglucose isomerase affects leaf starch synthesis and floral initiation 1. Plant Physiol 123:319–325
Yun MS, Umemoto T, Kawagoe Y (2011) Rice debranching enzyme isoamylase3 facilitates starch metabolism and affects plastid morphogenesis. Plant Cell Physiol 52:1068–1082. https://doi.org/10.1093/pcp/pcr058
Zhan Y, Li H, Sui M, et al (2020) Genome wide association mapping for tocopherol concentration in soybean seeds across multiple environments. Ind Crops Prod 154:112674. https://doi.org/10.1016/j.indcrop.2020.112674
Zhang H, Cui W (2006) Research progress in the factor affecting the content of starch in flue!cured tobacco leaf. J Anhui Agri Sci 34:6530–6533
Zhang X, Myers AM, James MG (2005) Mutations affecting starch synthase III in Arabidopsis alter leaf starch structure and increase the rate of starch synthesis. Plant Physiol 138:663–674. https://doi.org/10.1104/pp.105.060319
Zhong R, Teng Q, Haghighat M et al (2017) Cytosol-localized UDP-xylose synthases provide the major source of UDP-xylose for the biosynthesis of xylan and xyloglucan. Plant Cell Physiol 58:156–174. https://doi.org/10.1093/pcp/pcw179
Zhou Z, Jiang Y, Wang Z et al (2015) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol 33:408–414. https://doi.org/10.1038/nbt.3096
Funding
This work was supported by Tobacco Genome Projects of CNTC (Grant No. 110201901020 (JY-07))) and Science and Technology Projects of Henan Tobacco Company of CNTC (2021410000240021).
Author information
Authors and Affiliations
Contributions
XX and JY conceived and designed the research. XX, ZW, JFZ, XDX, and MZW performed the experiments. JFZ and ZPL contributed to genotyping. SXX, MX, and LH contributed to phenotypic evaluation. XX analyzed the data and wrote the manuscripts. ZW and JY revised the manuscript. All authors contributed to and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10142_2022_851_MOESM1_ESM.pptx
Supplementary file1 (PPTX 103 KB) Fig. S1 The qRT-PCR analysis for 10 candidate genes in parent lines with different allelic variants. a. Effect of two allelic variants ‘A’ and ‘G’ for Ntab0823160 on relative expression level for eight parent lines. b. Effect of two allelic variants ‘C’ and ‘T’ for Ntab0984470 on relative expression level for eight parent lines.c. Effect of two allelic variants ‘C’ and ‘T’ for Ntab0375050 on relative expression level for eight parent lines. d. Effect of two allelic variants ‘C’ and ‘T’ for Ntab0984590 on relative expression level for eight parent lines.e. Effect of two allelic variants ‘T’ and ‘G’ for Ntab0941870 on relative expression level for eight parent lines.f. Effect of two allelic variants ‘G’ and ‘A’ for Ntab0009730 on relative expression level for eight parent lines.g. Effect of two allelic variants ‘A’ and ‘C’ for Ntab0009530 on relative expression level for eight parent lines.h. Effect of two allelic variants ‘C’ and ‘T’ for Ntab0941490 on relative expression level for eight parent lines.i. Effect of two allelic variants ‘A’ and ‘G’ for Ntab0984540 on relative expression level for eight parent lines.j. Effect of two allelic variants ‘A’ and ‘G’ for Ntab0315480 on relative expression level for eight parent lines.
Rights and permissions
About this article
Cite this article
Xu, X., Wang, Z., Xu, S. et al. Identifying loci controlling total starch content of leaf in Nicotiana tabacum through genome-wide association study. Funct Integr Genomics 22, 537–552 (2022). https://doi.org/10.1007/s10142-022-00851-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-022-00851-x