Abstract
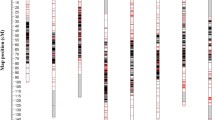
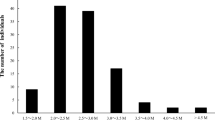
Genotyping-by-sequencing (GBS) allows rapid identification of markers for use in development of linkage maps, which expedite efficient breeding programs. In the present study, we have utilized GBS approach to identify and genotype single-nucleotide polymorphism (SNP) markers in an inter-specific RIL population of Cicer arietinum L. X C. reticulatum. A total of 141,639 raw SNPs were identified using the TASSEL-GBS pipeline. After stringent filtering, 8208 candidate SNPs were identified of which ~ 37% were localized in the intragenic regions followed by genic regions (~ 30%) and intergenic regions (~ 27%). We then utilized 6920 stringent selected SNPs from present study and 6714 SNPs and microsatellite markers available from previous studies for construction of linkage map. The resulting high-density linkage map comprising of eight linkage groups contained 13,590 markers which spanned 1299.14 cM of map length with an average marker density of 0.095 cM. Further, the derived linkage map was used to improve the available assembly of desi chickpea genome by anchoring 443 previously unplaced scaffolds onto eight linkage groups. The present efforts have refined anchoring of the desi chickpea genome assembly to 55.57% of the ~ 520 Mb of assembled desi genome. To the best of our knowledge, the linkage map generated in the present study represents one of the most dense linkage map developed for the crop till date. It will serve as a valuable resource for fine mapping and positional cloning of important quantitative trait loci (QTLs) associated with agronomical traits and also for anchoring and ordering of future genome sequence assemblies.






Similar content being viewed by others
References
Amalraj A, Taylor J, Bithell S, Li Y, Moore K, Hobson K, Sutton T (2019) Mapping resistance to Phytophthora root rot identifies independent loci from cultivated (Cicer arietinum L.) and wild (Cicer echinospermum PH Davis) chickpea. Theor Appl Genet 132(4):1017–1033
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9(3):208–18
Basu U, Srivastava R, Bajaj D, Thakro V, Daware A, Malik N, Upadhyaya HD, Parida SK (2018) Genome-wide generation and genotyping of informative SNPs to scan molecular signatures for seed yield in chickpea. Sci Rep 8(1):13240
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635
Choudhary S, Gaur R, Gupta S, Bhatia S (2012) EST-derived genic molecular markers: development and utilization for generating an advanced transcript map of chickpea. Theor Appl Genet 124(8):1449–1462
Collard BC, Jahufer MZZ, Brouwer JB, Pang ECK (2005) An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 142(1-2):169–196
Deokar AA, Ramsay L, Sharpe AG, Diapari M, Sindhu A, Bett K, Warkentin TD, Tar’an B (2014) Genome wide SNP identification in chickpea for use in development of a high density genetic map and improvement of chickpea reference genome assembly. BMC Genomics 15(1):708
Deokar A, Sagi M, Tar’an B (2019) Genome-wide SNP discovery for development of high-density genetic map and QTL mapping of ascochyta blight resistance in chickpea (Cicer arietinum L.). Theor Appl Genet, 1-12
Deschamps S, Llaca V, May GD (2012) Genotyping-by-sequencing in plants. Biology 1(3):460–483
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6(5):e19379. https://doi.org/10.1371/journal.pone.0019379
Fierst JL (2015) Using linkage maps to correct and scaffold de novo genome assemblies: methods, challenges, and computational tools. Front Genet 6:220
Gan Y, Johnston AM, Knight JD, McDonald C, Stevenson C (2010) Nitrogen dynamics of chickpea: Effects of cultivar choice, N fertilization, Rhizobium inoculation, and cropping systems. Can J Plant Sci 90(5):655–666
Garrido-Cardenas JA, Mesa-Valle C, Manzano-Agugliaro F (2018) Trends in plant research using molecular markers. Planta 247(3):543–557
Gaur R, Azam S, Jeena G, Khan AW, Choudhary S, Jain M, Yadav G, Tyagi AK, Chattopadhyay D, Bhatia S (2012) High-throughput SNP discovery and genotyping for constructing a saturated linkage map of chickpea (Cicer arietinum L.). DNA Res 19(5):357–73
Gaur R, Jeena G, Shah N, Gupta S, Pradhan S, Tyagi AK, Jain M, Chattopadhyay D, Bhatia S (2015) High density linkage mapping of genomic and transcriptomic SNPs for synteny analysis and anchoring the genome sequence of chickpea. Sci Rep 5:13387
Iquira E, Humira S, François B (2015) Association mapping of QTLs for sclerotinia stem rot resistance in a collection of soybean plant introductions using a genotyping by sequencing (GBS) approach. BMC Plant Biol 15(1):5
Jaganathan D, Thudi M, Kale S, Azam S, Roorkiwal M, Gaur PM, Kishor PK, Nguyen H, Sutton T, Varshney RK (2015) Genotyping-by-sequencing based intra-specific genetic map refines a “QTL-hotspot” region for drought tolerance in chickpea. Mol Gen Genomics 290(2):559–571
Jain M, Misra G, Patel RK, Priya P, Jhanwar S, Khan AW, Shah N, Singh VK, Garg R, Jeena G, Yadav M (2013) A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J 74(5):715–29
Jha UC (2018) Current advances in chickpea genomics: applications and future perspectives. Plant Cell Rep 37(7):947–965
Jukanti AK, Gaur PM, Gowda CL, Chibbar RN (2012) Nutritional quality and health benefits of chickpea (Cicer arietinum L.): a review. Br J Nutr 108:S11–S26
Kale SM, Jaganathan D, Ruperao P, Chen C, Punna R, Kudapa H, Thudi M, Roorkiwal M, Katta MA, Doddamani D, Garg V (2015) Prioritization of candidate genes in “QTL-hotspot” region for drought tolerance in chickpea (Cicer arietinum L.). Sci Rep 5:15296
Kim C, Guo H, Kong W, Chandnani R, Shuang LS, Paterson AH (2016) Application of genotyping by sequencing technology to a variety of crop breeding programs. Plant Sci 242:14–22
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugenics 12:172–175
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19(9):1639–1645
Kujur A, Bajaj D, Upadhyaya HD, Das S, Ranjan R, Shree T, Saxena MS, Badoni S, Kumar V, Tripathi S, Gowda CLL (2015a) Employing genome-wide SNP discovery and genotyping strategy to extrapolate the natural allelic diversity and domestication patterns in chickpea. Front Plant Sci 6:162
Kujur A, Upadhyaya HD, Shree T, Bajaj D, Das S, Saxena MS, Badoni S, Kumar V, Tripathi S, Gowda CLL, Sharma S (2015b) Ultra-high density intra-specific genetic linkage maps accelerate identification of functionally relevant molecular tags governing important agronomic traits in chickpea. Sci Rep 5:9468
Kujur A, Upadhyaya HD, Bajaj D, Gowda CLL, Sharma S, Tyagi AK, Parida SK (2016) Identification of candidate genes and natural allelic variants for QTLs governing plant height in chickpea. Sci Rep 6:27968
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997
Li H, Vikram P, Singh RP, Kilian A, Carling J, Song J, Burgueno-Ferreira JA, Bhavani S, Huerta-Espino J, Payne T, Sehgal D (2015) A high density GBS map of bread wheat and its application for dissecting complex disease resistance traits. BMC Genomics 16(1):216
Li Y, Ruperao P, Batley J, Edwards D, Khan T, Colmer TD, Pang J, Siddique KH, Sutton T (2018) Investigating drought tolerance in chickpea using genome-wide association mapping and genomic selection based on whole-genome resequencing data. Front Plant Sci 9:190
Mannur DM, Babbar A, Thudi M, Sabbavarapu MM, Roorkiwal M, Sharanabasappa BY, Bansal VP, Jayalakshmi SK, Yadav SS, Rathore A, Chamarthi SK (2019) Super Annigeri 1 and improved JG 74: two Fusarium wilt-resistant introgression lines developed using marker-assisted backcrossing approach in chickpea (Cicer arietinum L.). Mol Breed 39(1):2
Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J, Barry K, Munoz-Amatriain M, Close TJ, Wise RP, Schulman AH, Himmelbach A (2013) Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J 76:718–727
Merga B, Haji J (2019) Economic importance of chickpea: production, value, and world trade. Cogent Food Agric, 1615718
Parween S, Nawaz K, Roy R, Pole AK, Suresh BV, Misra G, Jain M, Yadav G, Parida SK, Tyagi AK, Bhatia S, Chattopadhyay D (2015) An advanced draft genome assembly of a desi type chickpea (Cicer arietinum L.). Sci Rep 5(1). https://doi.org/10.1038/srep12806
Poland JA, Rife TW (2012) Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 5(3):92–102
Pootakham W, Ruang-Areerate P, Jomchai N, Sonthirod C, Sangsrakru D, Yoocha T, Theerawattanasuk K, Nirapathpongporn K, Romruensukharom P, Tragoonrung S, Tangphatsornruang S (2015) Construction of a high-density integrated genetic linkage map of rubber tree (Hevea brasiliensis) using genotyping-by-sequencing (GBS). Front Plant Sci 6:367
Pratap A, Chaturvedi SK, Tomar R, Rajan N, Malviya N, Thudi M, Saabale PR, Prajapati U, Varshney RK, Singh NP (2017) Marker-assisted introgression of resistance to fusarium wilt race 2 in Pusa 256, an elite cultivar of desi chickpea. Mol Gen Genomics 292(6):1237–1245
Ratnaparkhe MB, Tekeoglu M, Muehlbauer FJ (1998) Inter-simple-sequence-repeat (ISSR) polymorphisms are useful for finding markers associated with disease resistance gene clusters. Theor Appl Genet 97(4):515–519
Riaño-Pachón DM, Ruzicic S, Dreyer I, Mueller-Roeber B (2007) PlnTFDB: an integrative plant transcription factor database. BMC Bioinf 8(1):42
Roorkiwal M, Rathore A, Das RR, Singh MK, Jain A, Srinivasan S, Gaur PM, Chellapilla B, Tripathi S, Li Y, Hickey JM (2016) Genome-enabled prediction models for yield related traits in chickpea. Front Plant Sci 7:1666. https://doi.org/10.3389/fpls.2016.01666
Roorkiwal M, Jain A, Kale SM, Doddamani D, Chitikineni A, Thudi M, Varshney RK (2018) Development and evaluation of high-density Axiom® Cicer SNP Array for high-resolution genetic mapping and breeding applications in chickpea. Plant Biotechnol J 16(4):890–901
Scheben A, Batley J, Edwards D (2017) Genotyping-by-sequencing approaches to characterize crop genomes: choosing the right tool for the right application. Plant Biotechnol J 15(2):149–161
Sonah H, O'Donoughue L, Cober E, Rajcan I, Belzile F (2015) Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol J 13(2):211–221
Srivastava R, Singh M, Bajaj D, Parida SK (2016) A high-resolution InDel (insertion–deletion) markers-anchored consensus genetic map identifies major QTLs governing pod number and seed yield in chickpea. Front Plant Sci 7:1362
Srivastava R, Upadhyaya HD, Kumar R, Daware A, Basu U, Shimray PW, Tripathi S, Bharadwaj C, Tyagi AK, Parida SK (2017) A multiple QTL-Seq strategy delineates potential genomic loci governing flowering time in chickpea. Front Plant Sci 8:1105
Thudi M, Upadhyaya HD, Rathore A, Gaur PM, Krishnamurthy L, Roorkiwal M, Nayak SN, Chaturvedi SK, Basu PS, Gangarao NV, Fikre A (2014) Genetic dissection of drought and heat tolerance in chickpea through genome-wide and candidate gene-based association mapping approaches. PLoS One 9(5):e96758
Upadhyaya HD, Bajaj D, Narnoliya L, Das S, Kumar V, Gowda CLL, Sharma S, Tyagi AK, Parida SK (2016) Genome-wide scans for delineation of candidate genes regulating seed-protein content in chickpea. Front Plant Sci 7:302
Van Ooijen JW (2006) JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, Netherlands, 33
Van Os H, Stam P, Visser RG, Van Eck HJ (2005) RECORD: a novel method for ordering loci on a genetic linkage map. Theor Appl Genet 112(1):30–40
Varshney RK (2016) Exciting journey of 10 years from genomes to fields and markets: some success stories of genomics-assisted breeding in chickpea, pigeonpea and groundnut. Plant Sci 242:98–107
Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, Cannon S, Baek J, Rosen BD, Tar'an B, Millan T (2013a) Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol 31(3):240–246
Varshney RK, Gaur PM, Chamarthi SK, Krishnamurthy L, Tripathi S, Kashiwagi J, Samineni S, Singh VK, Thudi M, Jaganathan D (2013b) Fast-track introgression of “QTL-hotspot” for root traits and other drought tolerance traits in JG 11, an elite and leading variety of chickpea. Plant Genome, 6(3)
Varshney RK, Kudapa H, Pazhamala L, Chitikineni A, Thudi M, Bohra A, Gaur PM, Janila P, Fikre A, Kimurto P, Ellis N (2015) Translational genomics in agriculture: some examples in grain legumes. Crit Rev Plant Sci 34(1-3):169–194
Varshney RK, Thudi M, Roorkiwal M, He W, Upadhyaya HD, Yang W, Bajaj P, Cubry P, Rathore A, Jian J, Doddamani D (2019) Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits. Nat Genet 51(5):857–864
Verma S, Gupta S, Bandhiwal N, Kumar T, Bharadwaj C, Bhatia S (2015) High-density linkage map construction and mapping of seed trait QTLs in chickpea (Cicer arietinum L.) using Genotyping-by-Sequencing (GBS). Sci Rep 5:17512
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93(1):77–78
Voss-Fels K, Snowdon RJ (2016) Understanding and utilizing crop genome diversity via high-resolution genotyping. Plant Biotechnol J 14:1086–1094
Wang R, Gangola MP, Irvine C, Gaur PM, Båga M, Chibbar RN (2019) Co-localization of genomic regions associated with seed morphology and composition in a desi chickpea (Cicer arietinum L.) population varying in seed protein concentration. Theor Appl Genet, 1–19.
Winter P, Benko-Iseppon AM, Hüttel B, Ratnaparkhe M, Tullu A, Sonnante G, Pfaff T, Tekeoglu M, Santra D, Sant VJ, Rajesh PN (2000). A linkage map of the chickpea (Cicer arietinum L.) genome based on recombinant inbred lines from a C. arietinum× C. reticulatum cross: localization of resistance genes for fusarium wilt races 4 and 5. Theor Appl Genet 101(7):1155–63
Xu Y, Li P, Yang Z, Xu C (2017) Genetic mapping of quantitative trait loci in crops. Crop J 5(2):175–184
Yang Z, Chen Z, Peng Z, Yu Y, Liao M, Wei S (2017) Development of a high-density linkage map and mapping of the three-pistil gene (Pis1) in wheat using GBS markers. BMC Genomics 18(1):567
Acknowledgments
SV, SP and HA acknowledges the Department of Biotechnology, India. We thank Dr. Fred Muehlbauer, Washington State University, USA, for providing the inter-specific chickpea mapping population.
Funding
This work was financially supported by the Department of Biotechnology (DBT) under the Next Generation Challenge Programme on Chickpea Genomics (grant number BT/PR12919/AGR/02/676/2009).
Author information
Authors and Affiliations
Contributions
SB conceived and designed the experiments. RG, SV and SP performed lab experiments. RG, SP and HA performed bioinformatics analysis and analyze the data. RG drafted the manuscript. SV, HA and SB helped in data interpretation and participated in drafting and correcting the manuscript critically. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gaur, R., Verma, S., Pradhan, S. et al. A high-density SNP-based linkage map using genotyping-by-sequencing and its utilization for improved genome assembly of chickpea (Cicer arietinum L.). Funct Integr Genomics 20, 763–773 (2020). https://doi.org/10.1007/s10142-020-00751-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-020-00751-y