Abstract
Two dozen field-collected Bacillus and a dozen Bacillus spizizenii wild-type strains from strain collections were selected on the basis of their antagonistic properties against the Gram-positive strain Micrococcus luteus. Based on their genetic and antibiotic profiles, they were characterized (subtilin encoding spaS gene sequences, mass spectrometric, and quantitative-reversed phase liquid chromatographic analyses, as well as the presence of the lanthionine cyclase protein SpaC by western blotting), seven novel producers of the lanthipeptide subtilin. Phylogenetic analyses of the subtilin-producing wild-type strains based on their 16S rRNA sequences showed that all seven strains could be classified as B. spizizenii: The field-collected strains HS and N5, as well as strains DSM 618, 1087, 6395, 6405, and 8439 from the German Collection of Microorganisms and Cell Cultures. To the best of our knowledge, all B. spizizenii strains described so far are characterized by the fact that they can produce a lanthipeptide of the subtilin family. Both the lanthipeptide structures and the organization and sequences of the 16S rRNA-encoding genes suggest a subdivision of B. spizizenii into subspecies: The subtilin-producing B. spizizenii strains are distinctly different from the entianin-producing B. spizizenii typing strain TU-B-10 T (DSM 15029 T).
Similar content being viewed by others
Introduction
Gram-positive spore-forming bacteria of the genus Bacillus are among the most widespread bacteria worldwide; they can be found on the one hand in the soil, in fresh or seawater, as well as in the air (Ferrari et al. 1993; Moszer et al. 2002), on the other hand in the gastrointestinal tract of ruminants and humans (Hong et al. 2005; Cutting 2011). Some B. subtilis strains share a long history of safe use in fermented foods such as Nattō (B. subtilis natto, Japanese fermented soybeans) (Sun et al. 2016) and Doenjang (B. subtilis together with Aspergillus oryzae, Korean soybean paste Yue et al. 2021). For this reason, B. subtilis is generally classified as safe (GRAS). According to the currently accepted definition, probiotics are “live microorganisms which provide health benefits when consumed,” for example by improving or restoring the gut flora (Hill et al. 2014). Very recently, it has been shown that lanthipeptide producing commensal strains of the human gastrointestinal tract reduce vancomycin-resistant Enterococcus faecium (VRE) colonization and represent potential probiotic agents (Kim et al. 2018). Lanthipeptides are gene-encoded small peptides (19–38 amino acids in length) that possess the unusual bridge-forming sulfur-containing amino acids meso-lanthionine and 3-methyl-lanthionine (Freund and Jung 1992; Stein 2005; Chatterjee et al. 2005; Arnison 2013; Letzel et al. 2014).
B. subtilis as well as very closely related B. spizizenii strains are able to produce more than two dozen antibiotics with an amazing variety of structures (Stein 2005). Peptide antibiotics represent the predominant class, among them lanthipeptides and non-ribosomal biosynthesized lipopeptide antibiotics from the surfactin, iturin, and fengycin-class (Stein 2005; Zhao et al. 2018; Caulier et al. 2019). One of the one of the earliest described lanthipeptides is subtilin, a 32-amino-acid peptide produced by a Bacillus strain (see Fig. 1A for a structure representation) that has been originally described 1944 (Jansen and Hirschmann 1944). The subtilin producing strain was originally isolated 1911–1912 by Karl Kellerman (Kellerman et al. 1912) and deposited as B. subtilis strain No. 6633 into the American Type Culture Collection (ATCC) by the “Bureau of Plant Industry, Soils, and Agricultural Engineering of the United States Department of Agriculture” (Garibaldi and Feeney 1949); this strain was proposed as B. spizizenii by Nakamura et al. (1999) and promoted by Dunlap et al. (2020). Furthermore, several B. subtilis wild-type strains produce the macrocyclic sactipeptide subtilosin A (Fig. 1B; Zheng et al. 1999; Stein et al. 2004) with a series of unusual intramolecular thioether linkages (Marx et al. 2001; Kawulka et al. 2003; Stein 2020). The lipoheptapeptide lactone surfactin (Fig. 1C) is a powerful surfactant with potent antimicrobial activities of surfactin are based on its detergent-like action on biological membranes (Heerklotz und Seelig 2001; Carrillo et al. 2003). Surfactin is distinguished by its exceptional emulsifying, foaming, antiviral, antitumor, anti-mycoplasma, and hypocholesterolemic activities (Peypoux et al. 1999; Kaspar et al. 2019).
Structure of Bacillus spizizenii peptide antibiotics. A Structure of the 32 amino-acid lanthipeptide subtilin: The amino acid residues (one-letter-code) are circled and all posttranslational modified amino acid residues are colored. ΔA (blue), 2,3-didehydroalanine (dehydrated serine) at positions 5 and 32; ΔB (blue), 2,3-didehydrobutyrine (dehydrated threonine) at position 18. The five ring structures represent intramolecular thioether bridges, namely the amino acids meso-lanthionine (A-s-A, linking amino acids 3 and 7) or methyllanthionine (Ab-s-A linking amino acids 8 and 11, 13 and 19, 23 and 26, as well as 25 and 28). The posttranslational modified cysteine residue is shown in yellow. B Structure of the cyclic 35 amino-acid sactipeptide subtilosin: The amino acid residues (one-letter-code) are circled and the posttranslational modified amino acid residues are colored, namely three cross-links between the sulfurs of Cys4, 7, and 13 (yellow) which are linked to the alpha-position of Phe22, Thr28, and Phe31 (pink), respectively (Kawulka et al. 2003). The N-and C-termini (green, positions 1 and 35, respectively) are linked via amide bond forming a macrolactam. C Structure of the lipoheptapeptide surfactin: Surfactin is a heptapeptide lactone, which consist of a ß-hydroxy fatty acid (purple) whose carboxyl group is linked to a heptapeptide moiety via amide bond. Lactonization is accomplished by esterification of the carboxy group of the C-terminal leucine (green) with the ß-hydroxy fatty acid. Further, non-proteinogenic components are the D-configured amino acids in positions 3 and 6 (orange). Variations with both the chain length (13–15 carbon atoms) and the branching pattern (n, iso, anteiso) of the fatty acid moiety, as well as the identity of the amino acid in position 7 (exchange from L-Leu to L-Ile or L-Val) lead to a microheterogeneity of natural surfactin produced by B. subtilis (Kowall et al. 1998)
The aim of this study was to analyze the potential of Bacillus strains to produce the lanthipeptide subtilin, the sactipeptide subtilosin, and the lipopeptide surfactin. Both laboratory and field collected Bacillus strains were taxonomically classified by gene sequencing (16SrRNA encoding rrn genes and subtilin encoding spaS gene). The antibiotic profiles were characterized under conditions optimal for the production of lanthi- and sactipeptides; MALDI mass spectrometry and reversed phase rpHPLC were used for qualitative and quantitative verification of the produced peptides. Furthermore, the presence of subtilin-modifying enzymes (SpaC, the subtilin cyclase, for example) was verified by immunoblotting.
Materials and methods
Bacterial strains and media
Bacillus strains (for a complete list see Table 1) from culture collections are from ATCC (American Type Culture Collection) and DSMZ (German Collection of Microorganisms and Cell Cultures; strain collections for microorganisms and links to them can be found in the Tab. S1, Supplement) or natural isolates (see below). For antibiotic production liquid Landy medium cultures were routinely grown aerobically at 37 °C (Landy et al. 1948) supplemented with 0.5% yeast extract (Heinzmann et al. 2006; Stein 2020). Solid media contained TY agar (0.8% tryptone, 0.5% yeast extract, 0.5% NaCl, 1.5% agar) or lysogeny broth (LB) agar (the commercial supplier of the media components was Gibco, Neu-Isenburg, Germany), standard incubation conditions were overnight (15–20 h) at 37 °C.
Isolation of subtilin producing Bacilli
Soil samples with different nutrient content were isolated from nutrient-rich arable land (N, 50° 11′ 1′′, N; 08° 47′ 54′′ E) and forest (IP, 50° 08′ 22′′ N; 09° 16′ 33′′ E) in Hessen, Germany, and from alpine surroundings (HI, 47° 21′ 13′′ N; 10° 06′ 01′′ E and HS, 47° 19′ 36′′ N; 10° 11′ 02′′ E; both approx. 2000 m, Vorarlberg, Austria). Twenty-five grams of soil were suspended in 100 mL of sterile water, pasteurized (10 min at 80 °C) in order to accumulate spore-forming bacteria, diluted 1: 100, plated out on TY plates and incubated at 37 °C for 15 h. To monitor the antimicrobial activities, individual colonies were replica plated on TY agar plates in Petri dishes with cams in order to obtain optimal aerobic conditions for antibiotic production (Sarstedt, Nümbrecht, Germany) with Micrococcus luteus ATCC 9341 as the test organism (Stein et al. 2004).
Molecular biology techniques
Established protocols were followed for molecular biology techniques; E. coli plasmids were isolated by the rapid alkaline extraction procedure. DNA amplification using Taq DNA polymerase was performed according to the instruction of the commercial supplier (Boehringer GmbH; Mannheim, Germany) in a DNA Thermal cycler (Eppendorf; Hamburg, Germany). The 16S rRNA encoding genes of the bacterial isolates were PCR amplified using primers 16S_forward 5′-GAGAGTTTGATCCTGGCTCAG-3′ and 16S_reverse 5′-ACGACTTCACCCCAATCATC-3′ (Heyrman et al. 2001). The 16S rRNA sequences of the Bacillus strains DSM 6405, 3258, N5, HS1, IP, and HI were deposited under the NCBI GeneBank records DQ452508-13, respectively; the DSM618 sequence under record DQ529249 (for NCBI GeneBank links see Tab. S4, Supporting Informations). The structural gene of subtilin spaS including 250 bp of the upstream non-coding region has been PCR-amplified with primers SpaS_Seq1 (5′-CTATGAATCAATGGAAGGG-3′) and SpaS_Seq2 (5′-CTTCATTTTCTTGTCCCG-3′); GeneBank records for B. spizizenii DSM 618 (DQ452514: https://www.ncbi.nlm.nih.gov/nuccore/DQ452514), B. spizizenii DSM 6405 (DQ452515: https://www.ncbi.nlm.nih.gov/nuccore/DQ452515), HS (DQ452516: https://www.ncbi.nlm.nih.gov/nuccore/DQ452516), and N5 (DQ452517:https://www.ncbi.nlm.nih.gov/nuccore/DQ452517). Subtilosin structural gene sbo sequencing has been performed with primers described previously (Stein 2004); GeneBank records for novel sbo sequences of the natural isolates B. subtilis HI-1, N5, and IP are deposited under accession numbers DQ452518-20, respectively. DNA cleavage and isolation were achieved with the QIAquick™ purification kit (Qiagen GmbH; Hilden, Germany). Oligonucleotides were purchased from ARK (ARK Scientific GmbH Biosystems, Darmstadt, Germany). Sequencing was carried out by SRD (Scientific Research and Development, Oberursel, Germany); nucleotide sequences have determined at least two times for each DNA-strand.
DNA sequence and phylogenetic analyses
The 16S rRNA sequences of B. subtilis strains 168 and ATCC 6051 were taken from the NCBI gene bank reference sequences NC_000964.3 and NZ_CP020102.1, respectively. The 16S rRNA sequences of the B. spizizenii strains ATCC 6633, W23, and TU-B-10 T were taken from NZ_CP034943.1, CP002183.1, and CP002905.1, respectively. GC content calculation and GC profiling of different B. subtilis genomes and subtilin gene clusters was performed using ENDMEMO (http://www.endmemo.com/bio/gc.php). Multiple sequence alignment (MSA) analyses were performed with Clustal Omega 1.2.4 that uses seeded guide trees and HMM profile-profile techniques to generate alignments (Waterhouse et al. 2009; Sievers et al. 2011) (https://www.ebi.ac.uk/Tools/msa/clustalo/) or MAFFT version 7 (Katoh et al. 2019; https://mafft.cbrc.jp/alignment/server/). Phylogenetic analyses were carried out by the neighbor joining method (Saitou and Nei 1987) using MAFFT (version 7; Kuraku et al. 2013) or Clustal Omega 1.2.4 software. The NCBI BLAST server (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for homology searches.
MALDI-TOF mass spectrometry (MALDI-TOF MS)
Cell-free aliquots of a 500-µL culture supernatants of overnight grown Bacillus strains in Landy media were extracted with 500 µL 1-butanol, 400 µL of the organic phase was dried in a speed-vac evaporator, and the extracted peptides were dissolved in 5 µL 50% acetonitrile and 1% trifluoroacetic acid (v/v in H2O). 0.3–0.5 µL aliquots of the solutions were mixed directly on the target with 1.5 µL matrix solution, and the mixture was dried with the help of hot air. The matrix was 20 mg/mL DHBs (9:1 mixture of 2,5-dihydroxybenzoic acid and 2-hydroxy-5-methoxybenzoic acid) complemented with solubilized in aqueous solution of 50% acetonitrile and 1% trifluoroacetic acid (v/v in H2O). Generally, if sample spots are readily crystallized, mass spectra with sufficient signal-to-noise values were obtained from the edge of the crystals (Stein 2008). Delayed extraction™ (DE) MALDI time-of-flight (TOF) mass spectra were recorded on a Voyager-DE STR instrument (Applied Biosystems Instruments) using a nitrogen laser (λ=336 nm, repetition rate = 20 Hz) for desorption and ionization with an acquisition mass range from 600 to 15 000 m/z and the low mass gate set to 550 m/z. The total acceleration voltage was 20 kV with 68.5% grid voltage on the first grid, 0.02% guide wire voltage, 200 ns delay, and a mirror voltage ratio of 1.12. All experiments were carried out with the reflector positive ion mode. Between 500 and 1 000 laser shots were accumulated for each mass spectrum.
Subtilin quantitation
Similar growth profiles were obtained for all B. spizizenii strains tested after growth in 2 mL Landy medium (37 °C, 175 rpm). In standard determinations, B. spizizenii was grown for 16 h, the cultures were centrifuged (4 °C, 15 min, 15,000 g), and the subtilin amount was determined in 500-µL aliquots by RP-HPLC using a Beckman Gold HPLC System and an analytical ODS Hypersil column (particle size: 5 µm, width and length: 2 × 250 mm, Maisch, Ammerbuch, Germany) as described previously (Heinzmann et al. 2006).
SDS-PAGE and Western blotting
SDS-PAGE (10% tris–glycine gels) and Western blot analyses were performed with an immuno-purified SpaC-directed immunoserum as described previously (Helfrich et al 2007). Molecular standards were purchased from BioRad (München, Germany).
Results and discussion
Identification of antibiotic producer among field collected spore forming bacteria
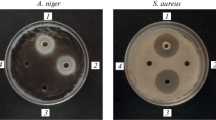
We took random soil samples from various environmental habitats, nutrient-rich farmland, forest (200-m altitude, Hesse, Germany), as well as alpine environments (2000 m altitude, Vorarlberg, Austria). After pasteurization (10 min, 80 °C), spore-forming bacteria were plated on agar plates containing TY medium. Single colonies were selected and examined for their antibiotic activities against M. luteus as a highly sensitive Gram-positive target strain using optimized conditions for the detection of lanthipeptides (Heinzmann et al. 2006) and sactipeptides (Stein et al. 2002a, b). Twenty to 25% of the isolated aerobically grown spore-formers significantly inhibited M. luteus growth. Twelve potential antibiotic producers were identified: six strains from farmland (N5), one from a mixed forest (IP), and five strains from alpine environments (HS and HI). Strains HS and N5 exhibited large and clear zones of inhibition comparable to the established subtilin-producer B. spizizenii ATCC 6633 (Fig. 2A) and were later characterized as subtilin producers (Sub+, for a summary see Table 1). In contrast, strains HI and IP exhibited inhibition zones that were only about one quarter the size of those of B. spizizenii ATCC 6633). Both strains were classified as Sub− in further studies (for a summary see Tab.1). Additionally, we tested twelve B. subtilis and B. spizizenii wild-type from the German Collection of Microorganisms (DSMZ) for their ability to inhibit M. luteus growth. Strains DSM 618, 1087, 6395, 6405, and 8439 showed promising antimicrobial activities with growth inhibition activities comparable to the well-characterized subtilin-producer B. spizizenii ATCC 6633; representative examples are given in Fig. 2A. The B. subtilis strains DSM 3256 and DSM 3258 exhibited semi-large inhibition zones (Fig. 2A).
Identification of subtilin producing Bacillus strains. Representative examples of wild-type Bacillus spizizenii strains DSM 618, DSM 3258, and DSM 6405; the natural isolates are designated HS, N5, HI, and IP. The references for subtilin producers and nonproducers were the strains B. spizizenii ATCC 6633 (Sub+) and B. subtilis 168 (Sub−), respectively. A Analyses of Bacillus strains in agar diffusion tests using M. luteus as the test organism. The production of antimicrobial agents is indicated by growth inhibition halos of the test organism. For B. subtilis 168 no growth inhibition halo was detected. Large growth inhibition halos comparable to the Sub+ strain B. spizizenii 6633 were obtained for the strains HS, N5, DSM 6018, and DSM 6405. The halos for the strains DSM 3258 as well as for HI and IP were significantly smaller. B The subtilin structural gene spaS gene was PCR amplified using the primers SpaS_Seq1 and 2, as well as DNA from different Bacillus cells as template. The right lane contains a nucleotide size standard. The calculated size of the amplified DNA segment containing spaS is 480 bp. The subtilin gene spaS was identified in strains with large inhibition halos (B. spizizenii, HS, N5, DSM 6018, and DSM 6405), whereas it was not detected in strains with intermediate inhibition halos. C Western blot analyses: Bacillus cells harvested in the stationary growth phase were lysed. After SDS-PAGE separation of cell lysates, the proteins were transferred nitrocellulose membranes and the presence of the SpaC protein was analyzed with SpaC-directed immunoserum. Consistent with the results shown in Fig. 2B, the SpaC protein (calculated molar mass of 42,5 kDa) was found in all strains containing the subtilin gene spaS
Phylogenetic classification of Bacillus strains on the basis of 16S rRNA sequences
The 16S rRNA encoding genes of all field-collected spore-forming strains were PCR amplified and sequenced. Remarkably, the 16S rRNA sequences of the field-collected Bacillus strains HS1 and HS2 (hereinafter referred to as strain HS) and N1, N5, and N6 (referred to as strain N5), as well as the DSMZ strains DSM 618 and DSM 6405 were identical to the database-established 16S rRNA sequences of B. spizizenii ATCC 6633 and W23 (Fig. 3A). This suggests their classification as B. spizizenii, a species first postulated by Nakamura et al. (1999) and promoted by Dunlap et al. (2020). As indicated in Fig. 3B, MSA analyses revealed that position 181 of the individual 16S rRNA encoding rrn genes can be used for clear species differentiation between B. spizizenii (C at position 181) and B. subtilis (G at position 181 in all ten rrn genes of B. subtilis 168 and ATCC 6051).
16S rRNA sequence analysis of different Bacillus strains. A Multiple sequence alignment: The sequences of the ten 16S rRNA encoding rrn genes of a given. B. subtilis strain are almost identical (99.81%; see also Tab. S4, supporting information). Two representative 16S rRNA sequences are shown for B. subtilis strains 168 and ATCC 6051 (rrnO and rrnJ genes). Since the 16S rRNA sequences within B. spizizenii strains ATCC 6633, W23, and TUB are almost identical, only one representative rrn sequence is shown. All 16S rRNA genes of the newly identified subtilin-producing strains exhibited 100% identity to the B. spizizenii reference strains ATCC 6633 and W23. The positions 181 and 600 of the 16S rRNA sequences clearly differentiated the Bacillus strains B. subtilis and B. spizizenii (red box). B Neighbor joining phylogenetic tree: The strains DSM 3258, HI, and IP were classified as B. subtilis (blue bracket) based on the similarity of their 16S rRNA sequences to those of B. subtilis 168 and ATCC 6051
On the other hand, the field-collected strains IP and HI1 (referred to as strain HI) as well as the laboratory strain DSM 3258 are classified as further members of B. subtilis (see Fig. 3B for phylogenetic analyses). 16S rDNA-based classification revealed that the field collected Bacillus strains HI2 and HI3 belong to B. macroides and B. licheniformis, respectively. Both species are known for members with a high potential to produce antibiotics. The strain N2 was classified as B. thuringiensis, a class of Bacillus strains some members of which produce the lipopeptide kurstakin (Hathout et al. 2000). No sequence was obtained for strain N3, and strain N4 was discarded because no further significant antibiotic activities could be identified under the test conditions used.
Diagnostic PCR and DNA sequencing of the subtilin structural gene spaS
For PCR amplification of the subtilin structural gene spaS oligonucleotide primers (SpaS-Seq1 and SpaS_Seq2) complementary to the –35 region of the spaS-promoter and the spaS-spaI intergenic region were used. The presence of the spaS gene was verified in the case of B. spizizenii strains DSM 618 and 6405, as well as the natural isolates HS and N5 (Fig. 2B; for a summary see Table 1). The corresponding spaS genes and their flanking regions (ribosomal binding site, -10-region) were sequenced (the GeneBank records for B. spizizenii DSM 618, 6405, HS1, and N5 are DQ452514-17, respectively). The spaS sequences were identical to the spaS sequence of the B. spizizenii strain ATCC 6633 strain ATCC 6633: NZ_CP034943.1). For the B. spizizenii strains containing the spaS gene also, the subtilin cyclase SpaC (Fig. 2C) was detected within PAGE-separated cell extracts using SpaC specific immunosera. Most likely, all observed SpaC proteins of the B. spizizenii strains are closely related, since immunoblot signals of potential subtilin producers showed comparable intensities (Fig. 2C). In contrast, an immunoblot of EriC from B. subtilis A1/3, a protein exhibiting only 85% sequence identity with the SpaC counterpart from B. spizizenii ATCC 6633 (Stein et al. 2002a), showed only very weak immunoblotting signals. Consistently, for the strains which lack the subtilin structural gene spaS (strains 168, 3258, HI, and IP), no SpaC protein could be detected in the associated cell extracts either (Fig. 2B).
Identification of subtilin within B. spizizenii culture supernatants by MALDI-TOFMS
MALDI-TOFMS analyses of butanolic extracts of B. spizizenii culture supernatants resulted in prominent peak cluster between m/z 3280 and 3520 (Fig. 4). They represent H+-, Na+-, and K+-adducts of the lanthipeptide subtilin and its succinylated derivative (Chan et al. 1993; Heinzmann et al. 2006), as well as the sactipeptide subtilosin, respectively (see Table 1 for summary). Furthermore, the lipoheptapeptide surfactin was identified by MALDI-MS experiments due to the characteristic m/z values of its different isoforms (Fig. 4). Remarkably, all investigated B. subtilis and B. spizizenii strains produced surfactin (see Table 1 for a summary), notably, with the exception of the laboratory-adapted B. subtilis strain 168. For this strain, a mutation within the 4′-phosphopantetheine transferase sfp gene which posttranslationally modifies the required surfactin synthetase enzymes so that surfactin cannot be produced (Lambalot et al. 1996). However, the detection of surfactin within the culture supernatants of all investigated B. subtilis and B. spizizenii strains, and the widespread frequent appearance of surfactin producers among strains of the genus Bacillus (Peypoux et al. 1999; Kalinovskaya et al. 2002; Torres et al. 2016) restricts the usage of the phenotype “surfactin production” as biomarker for subspecies classification/differentiation (e.g., between B. subtilis and B. spizizenii).
MALDI-TOF MS of a butanolic extract of a representative B. spizizenii (DSM 618) culture supernatant: The mass spectrum was recorded in the positive ion modus. Left insert, positively charged sodium (m/z 1030.7, 1044.7, 1058.7, and 1072.7) and potassium (1046.7, 1060.7, 1074.7, and 1088.7) adducts of different isoforms of the lipopeptide isoform surfactin A are labeled. The number of carbon atoms within the surfactin acyl chain is denoted in the index of SrfA (Kowall et al. 1998). In the right insert, the proton-adducts of subtilin (Sub, m/z 3322.8) and subtilosin (Sbo, 3400.8) are labeled as well as their sodium- and potassium adducts (Sub, 3344.8, 3360.9; Sbo, 3438.9). SucSub represents the potassium-adduct of succinylated subtilin at m/z 3460.9; the peak at m/z 3422.8 can be interpreted as [SucSub + H]+ or [Sbo + Na].+ (asterix)
Quantitative determination of subtilin
The subtilin concentration was determined in Landy-culture supernatants of stationary grown Bacillus cells (Fig. 5A/B). Whereas the production yields of B. spizizenii ATCC 6633 (4.9 mg/mL) and the field-collected strain N5 (4.2 mg/mL) were comparable, the strain DSM 618 produces three-fold higher amounts (14.9 mg/mL). The largest subtilin yield was obtained from B. spizizenii DSM 6405 (33 mg/mL) and the field-collected B. spizizenii HS (30 mg/mL). Representative chromatograms for these strains are shown in Fig. 5A. The production yields of the DSM strains 1087, 6395, and 8439 were similar to the ATCC 6633 strain. Our finding that different strains produce different amounts of subtilin––the yield of the HS strain was sevenfold superior to the yield obtained from the original subtilin producer B. spizizenii ATCC 6633 (Heinzmann et al. 2006) and imply differential efficiencies in subtilin production. The examined B. spizizenii strains may have developed different genetic elements for the regulation of the extremely complex system of subtilin biosynthesis, such as repressor (AbrB) or activator (Sigma factor H) elements or variations in the promoter regions (-35 regions) (Stein et al. 2002b and 2003; Kleerebezem 2004; Kleerebezem et al. 2004; Spieß et al. 2015; Zhang et al, 2022). Furthermore, also the subsequent steps in subtilin biosynthesis for example post-translational dehydration of serine (threonine), addition of neighboring cysteines (Kiesau et al. 1997; Helfrich et al. 2007), and final processing (Stein and Entian 2002; Corvey et al. 2003) might exhibit differential efficiency in the investigated strains.
Subtilin production by different B. spizizenii strains. A Identification of subtilin by quantitative RP-HPLC. B. spizizenii strains were grown for 16 h in Landy medium to comparable cell densities; the strain B. subtilis 168 was used as a non-subtilin producing control (dotted line in black). Five hundred microliters of aliquots of cell-free culture supernatants from stationary cells were separated by reverse phase HPLC. Antimicrobial growth assays and MALDI-TOF–MS experiments indicated that subtilin elutes as a single peak at 17.5 to 18.5 min. B Quantitative determination of subtilin produced by different B. spizizenii strains. The integrals of the peaks eluting at 17.5–18.5 min (Fig. 6A) are proportional to the amount of subtilin. The presented values with standard errors of less than ± 12% are the means of three independent cultures for which the determinations have been performed twice
GC content of the subtilin gene cluster spa
Our results show that for all B. spizizenii strains characterized so far, the Sub+ phenotype is a characteristic feature. The analysis of the base compositions of a given genome is a common strategy to investigate gene history (Garcia-Vallvé et al. 2000; Popa et al. 2001). Remarkably, in all analyzed B. spizizenii genomes, the average GC content of the subtilin gene cluster of 36% is significantly lower (about 8%) than the average GC content of the respective B. spizizenii host genome of about 44% (Fig. 6 and Tab. S3, supporting information). This observation is a strong hint that B. spizizenii acquired the subtilin gene cluster most likely from another microorganism by a recent horizontal gene transfer event as is hypothesized for a number of lanthipeptide producers (Zhang et al. 2012).
Comparison of the GC content of the subtilin gene cluster and its flanking regions with the GC content of the B. subtilis and B. spizizenii genomes. A The GC content of the 12 kb subtilin gene cluster (red balls) of B. spizizenii (W23 and ATCC 6633), the upstream localized opuB gene cluster (green), and the downstream localized gene region of subtilin non-producers (black) were plotted versus the GC-content of the cognate genomes (black horizontal dotted lines). B GC content of the spa gene cluster of B. spizizenii ATCC 6633 and its flanking regions (inverse orientation). Insert: The subtilin cluster of B. spizizenii ATCC 6633 encoding the subtilin gene cluster spaBCSFEGRK (11.8 kbp, nucleotide positions 3,251,415–3,263,272, negative strand). Red, spaS encoding the subtilin pre-propeptide; blue the posttranslational modification machinery spaBC and the transporter spaT; purple, genes spaIFEG involved in immunity against matured subtilin; orange, spaRK encoding a two component regulatory system (Stein 2005). Black arrows represent different polycistronic transcripts of the spa-cluster. Upstream of the spa gene cluster is the opuB-operon (green), which is involved in choline transport (Hofmann and Brenner 2017); catR downstream of the spa cluster belongs to the MarR/DUF24 family of transcription repressors (Chi et al. 2010)
The subtilin producing B. spizizenii strains differ from the B. spizizenii typing strain TU-B-10 T in 16S rDNA and lanthipeptide sequence
Nakamura et al. (1999) proposed the classification of B. subtilis strains into two classes: (1) The 168-type strains into B. subtilis subsp. subtilis, and (2) the W23 strains (Zeigler et al. 2008; Zeigler, 2011) into B. subtilis subsp. spizizenii. Classical chemotaxonomy differentiates between both classes by the composition of their cell wall teichoic acids, whereas 168-type strains are endowed with the essential major teichoic acids poly(glycerol phosphate) and the non-essential minor teichoic acids poly(glucopyranosyl N-acetylgalactosamine 1-phosphate), the W-23-type mainly consists of poly(ribitol phosphate) (Lazarevic et al. 2002). Very recently, B. subtilis subsp. spizizenii was promoted to species status on the basis of comparative genomics and secondary metabolite (mycosubtilin and bacillaene) production (Dunlap et al. 2020).
The experiments presented in this study have revealed that all B. spizizenii strains produced the lanthipeptide subtilin. Surprisingly, a comparison of both, 16S rDNA gene organization and 16S rDNA gene sequences showed that the subtilin producing B. spizizenii strains characteristically differ from the B. spizizenii typing strain TU-B-10 T (DSM 15029 T). B. spizizenii ATCC 6633 has ten 16S rRNA encoding genes (rrn) identical in length and sequence, whereas only eight rrn genes were found for its close relative, the strain W23 (Table S2, Supplement). The B. spizizenii typing strain TU-B-10 T contains ten 16S rDNA genes (rrn genes) with a different genetic arrangement than the subtilin producing B. spizizenii strains: The first five rrn genes (rrnO, A, J, W, and I) are arranged similarly to the rrn gene organization of B. spizizenii ATCC 6633 and even B. subtilis 168 (Tab. S2 Supplement). However, the rrn genes 6–10 are located at different positions in the genome than those of B. spizizenii ATCC 6633 and B. subtilis 168 indicating genetic restructuring. The 16S rDNA encoding sequence of the subtilin producing B. spizizenii strains ATCC 6633, DSM 618, DSM 6405, N5, and HS, have a C, T and C at positions 181, 279, and 600, respectively (Fig. 3A). These three positions can be used to clearly differentiate subtilin producing B. spizizenii from subtilin non-producing B. subtilis strains (G, C/T, T at positions 181, 279, and 600, respectively). Furthermore, these positions can be even used to differentiate the subtilin producer from the B. spizizenii typing strain TU-B-10 T: C, T, T at position 181, 279, and 600, respectively (Fig. 3A). In this context, it is important that for the TU-B-10 T strain, the production of a lanthipeptide (entianin) was described that differs from subtilin in three amino acids: L6V, A15L, and L24I (Fuchs et al. 2011). Taken together, the differences in both the lanthipeptide structures and the organization and sequences of the 16S rRNA-encoding genes suggest the split of B. spizizenii into subspecies (Fig. 3B): the entianin producing B. spizizenii typing strain TU-B-10 T and the subtilin producing novel B. spizizenii subspecies strains.
Conclusion
In an age in which bacterial resistance to antibiotics is becoming increasingly important, systematic screening for new antibiotic agents with new mechanisms of action is still an important strategy. Microorganisms from the genus Bacillus are able to produce a large number of different antimicrobial substances (Stein 2005; Zhao et al. 2018; Tran et al. 2022). It is to be expected that a large number of new active antimicrobial agents and isoforms of known antibiotics with minor chemical modifications will be found in the future through systematic screening, a strategy which is strongly supported by subsequent genome sequencing and novel genome mining bioinformatic tools (Walker et al. 2020). The results of this work show that many B. spizizenii strains always produce a cocktail of antibiotic agents (Stein 2004 and 2020; Mülner et al. 2020), in particular several lipophilic membrane-active agents like subtilin, subtilosin, surfactin, and fengycin: In addition to classical 16S rDNA-based typing, determination of individual antibiotic cocktails (e.g., by mass spectrometric analyses) can contribute to very efficient subclassification of B. subtilis (Dunlap et al. 2020) and B. spizizenii (this study) strains.
References
Arnison PG, Bibb MJ, Bierbaum G, Bowers AA et al (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. https://doi.org/10.1039/C2NP20085F
Carrillo C, Teruel JA, Aranda FJ, Ortiz A (2003) Molecular mechanism of membrane permeabilization by the peptide antibiotic surfactin. Biochim Biophys Acta 1611:91–97. https://doi.org/10.1016/S0005-2736(03)00029-4
Caulier S, Nannan C, Gillis A, Licciardi F, Bragard C, Mahillon J (2019) Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front Microbiol 10:302. https://doi.org/10.3389/fmicb.2019.00302
Chan WC, Bycroft BW, Leyland ML, Lian LY, Roberts GC (1993) A novel post-translational modification of the peptide antibiotic subtilin: isolation and characterization of a natural variant from Bacillus subtilis A.T.C.C. 6633. Biochem J 291:23–27. https://doi.org/10.1042/bj2910023
Chatterjee C, Paul M, Xie L, van der Donk WA (2005) Biosynthesis and mode of action of lantibiotics. Chem Rev 105:633–684. https://doi.org/10.1021/cr030105v
Chi BK, Kobayashi K, Albrecht D, Hecker M, Antelmann H (2010) The paralogous MarR/DUF24-family repressors YodB and CatR control expression of the catechol dioxygenase CatE in Bacillus subtilis. J Bacteriol. 192:4571–4581. https://doi.org/10.1128/JB.00409-10
Corvey C, Stein T, Düsterhus S, Karas M, Entian K-D (2003) Activation of subtilin precursors by Bacillus subtilis extracellular serine proteases subtilisin (AprE), WprA, and Vpr. Biochem Biophys Res Commun 304:48–54. https://doi.org/10.1016/S0006-291X(03)00529-1
Cutting SM (2011) Bacillus probiotics. Food Microbiol 28:214–220. https://doi.org/10.1016/j.fm.2010.03.007
Dunlap CA, Bowman MJ, Zeigler DR (2020) Promotion of Bacillus subtilis subsp. inaquosorum, Bacillus subtilis subsp. spizizenii and Bacillus subtilis subsp. stercoris to species status. Antonie Van Leeuwenhoek 113:1–12. https://doi.org/10.1007/s10482-019-01354-9
Ferrari E, Jarnagin A, Schmidt BF (1993) Commercial production of extracellular enzymes. In: Sonenshein AL, Hoch JA, Losick R (eds) Bacillus subtilis and other Gram-positive bacteria. American Society for Microbiology, Washington, DC, pp 917–937
Freund S, Jung G. (1992) Lantibiotics: an overview and conformational studies on gallidermin and Pep5. In: James R, Lazdunski C, Pattus F. (eds) Bacteriocins, microcins and lantibiotics. NATO ASI Series (Series H: Cell Biology), vol 65. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-76974-0_9
Fritze D, Pukall R (2001) Reclassification of bioindicator strains Bacillus subtilis DSM 675 and Bacillus subtilis DSM 2277 as Bacillus atrophaeus. Int J Syst Evol Microbiol 51:35–37. https://doi.org/10.1099/00207713-51-1-35
Fuchs SW, Jaskolla TW, Bochmann S, Kötter P, Wichelhaus T, Karas M, Stein T, Entian K-D (2011) Entianin, a novel subtilin-like lantibiotic from Bacillus subtilis subsp. spizizenii DSM 15029T with high antimicrobial activity. Appl Environ Microbiol 77:1698–1707. https://doi.org/10.1128/aem.01962-10
Garcia-Vallvé S, Romeu A, Palau J (2000) Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res 10(11):1719–1725. https://doi.org/10.1101/gr.130000
Garibaldi JA, Feeney RF (1949) Subtilin production. Ind Eng Chem 41:432–435. https://doi.org/10.1021/ie50470a040
Hathout Y, Ho YP, Ryzhov V, Demirev P, Fenselau C (2000) Kurstakins: a new class of lipopeptides isolated from Bacillus thuringiensis. J Nat Prod 63:1492–1496. https://doi.org/10.1021/np000169q
Heerklotz H, Seelig J (2001) Detergent-like action of the antibiotic peptide surfactin on lipid membranes. Biophys J 81:1547–1554. https://doi.org/10.1016/S0006-3495(01)75808-0
Heinzmann S, Entian K-D, Stein T (2006) Engineering Bacillus subtilis ATCC 6633 for improved production of the lantibiotic subtilin. Appl Microbiol Biotechnol 69:532–536. https://doi.org/10.1007/s00253-005-0023-9
Helfrich M, Entian K-D, Stein T (2007) Structure-function relationships of the lanthionine cyclase SpaC involved in biosynthesis of the Bacillus subtilis peptide antibiotic subtilin. Biochemistry 46:3224–3233. https://doi.org/10.1021/bi062124f
Heyrman J, Swings J (2001) 16S rRNA sequence analysis of bacterial isolates from biodeteriorated mural paintings in the Servilia tomb (Necropolis of carmona, Seville, Spain). Syst Appl Microbiol 24:417–422. https://doi.org/10.1078/0723-2020-00048
Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME (2014) The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11:506–514. https://doi.org/10.1038/nrgastro.2014.66
Hofemeister J, Conrad B, Adler B, Hofemeister B, Feesche J, Kucheryava N, Steinborn G, Franke P, Grammel N, Zwintscher A, Leenders F, Hitzeroth G, Vater J (2004) Genetic analysis of the biosynthesis of non-ribosomal peptide- and polyketide-like antibiotics, iron uptake and biofilm formation by Bacillus subtilis A1/3. Mol Genet Genomics 272:363–378. https://doi.org/10.1007/s00438-004-1056-y
Hong HA, Le HD, Cutting SM (2005) The use of bacterial spore formers as probiotics. FEMS Microbiol Rev 29:813–835. https://doi.org/10.1016/j.femsre.2004.12.001
Jansen EF, Hirschmann DJ (1944) Subtilin - an antibacterial product of Bacillus subtilis: Culturing conditions and properties. Arch Biochem 4:297–309
Kalinovskaya NI, Kuznetsova TA, Ivanova EP, Romanenko LA, Voinov VG, Huth F, Laatsch H (2002) Characterization of surfactin-like cyclic depsipeptides synthesized by Bacillus pumilus from ascidian Halocynthiarantium. Mar Biotechnol (NY) 4:179–188. https://doi.org/10.1007/s10126-001-0084-4
Kaspar F, Neubauer P, Gimpel M (2019) Bioactive secondary metabolites from Bacillus subtilis: a comprehensive review. J Nat Prod 82:2038–2053. https://doi.org/10.1021/acs.jnatprod.9b00110
Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20:1160–1166. https://doi.org/10.1093/bib/bbx108
Kawulka K, Sprules T, McKay RT, Mercier P, Diaper CM, Zuber P, Vederas JC (2003) Structure of subtilosin A, an antimicrobial peptide from Bacillus subtilis with unusual posttranslational modifications linking cysteine sulfurs to alpha-carbons of phenylalanine and threonine. J Am Chem Soc 125:4726–4727. https://doi.org/10.1021/ja029654t
Kellerman KF, McBeth IG (1912) The fermentation of cellulose. Zentralblatt Für Bakteriologie, Parasitenkunde, Infektionskrankheiten Und Hygiene Abt II 34:485–494
Kiesau P, Eikmanns U, Gutowski-Eckel Z, Weber S, HammelmannM E-D (1997) Evidence for a multimeric subtilin synthetase complex. J Bacteriol 179:1475–1481. https://doi.org/10.1128/jb.179.5.1475-1481.1997
Kim J-Y, Park M-K, LeeY-J, Yi H, BahnY-S, Kim JF, Lee D-W (2018) The whole genome sequencing and assembly of Bacillus vallismortis DSM 11031T strain EMBL/GenBank/DDBJ databases. CP026362.1. Accessed 29 January 2022
Kleerebezem M (2004) Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 25:1405–1414. https://doi.org/10.1016/j.peptides.2003.10.021
Kleerebezem M, Bongers R, Rutten G, de Vos WM, Kuipers OP (2004) Autoregulation of subtilin biosynthesis in Bacillus subtilis: the role of the spa-box in subtilin-responsive promoters. Peptides 25:1415–1424. https://doi.org/10.1016/j.peptides.2003.11.025
Kowall M, Vater J, Kluge B, Stein T, Franke P, Ziessow D (1998) Separation and characterization of surfactin isoforms produced by Bacillus subtilis OKB 105. J Colloid Interface Sci 204:1–8. https://doi.org/10.1006/jcis.1998.5558
Kunst F, Ogasawara N, Moszer I et al (1997) The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249–256. https://doi.org/10.1038/36786
Kuraku S, Zmasek CM, Nishimura O, Katoh K (2013) aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acid Res. W22-W28. https://doi.org/10.1093/nar/gkt389
Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT (1996) A new enzyme superfamily - the phosphopantetheinyl transferases. Chem Biol 3:923–936. https://doi.org/10.1042/bj2910023
Landy M, Warren GH, Rosenman SB, Colio LG (1948) Bacillomycin: an antibiotic from Bacillus subtilis active against pathogenic fungi. Proc Soc Exp Biol Med 67:539–541. https://doi.org/10.3181/2F00379727-67-16367
Lazarevic V, Abellan FX, Möller SB, Karamata D, Mauël C (2002) Comparison of ribitol and glycerol teichoic acid genes in Bacillus subtilis W23 and 168: identical function, similar divergent organization, but different regulation. Microbiology 148:815–824. https://doi.org/10.1099/00221287-148-3-815
Leenders F, Stein T, Kablitz B, Franke P, Vater J (1999) Rapid typing of Bacillus subtilis strains by their secondary metabolites using matrix-assisted laser desorption/ionization mass spectrometry of intact cells. Rapid Commun Mass Spectrom 13:943–949. https://doi.org/10.1002/(SICI)1097-0231(19990530)13:10%3c943::AID-RCM591%3e3.0.CO;2-0
Letzel AC, Pidot SJ, Hertweck C (2014) Genome mining for ribosomally synthesized and post-translationally modified peptides (RiPPs) in anaerobic bacteria. BMC Genomics 15:983. https://doi.org/10.1186/1471-2164-15-983
Marx R, Stein T, Entian KD, Glaser SJ (2001) Structure of the Bacillus subtilis peptide antibiotic subtilosin A determined by 1H-NMR and matrix assisted laser desorption/ionization time-of-flight mass spectrometry. J Protein Chem 20:501–506. https://doi.org/10.1023/A:1012562631268
Moszer I, Jones LM, Moreira S, Fabry C, Danchin A (2002) SubtiList: the reference database for the Bacillus subtilis genome. Nucleic Acids Res 30:62–65. https://doi.org/10.1093/nar/30.1.62
Mülner P, Schwarz E, Dietel K, Junge H, Herfort S, Weydmann M, Lasch P, Cernava T, Berg G, Vater J (2020) Profiling for bioactive peptides and volatiles of plant growth promoting strains of the Bacillus subtilis complex of industrial relevance. Front Microbiol 11:1432. https://doi.org/10.3389/fmicb.2020.01432
Nakamura LK, Roberts MS, Cohan FM (1999) Relationship of Bacillus subtilis clades associated with strains 168 and W23: a proposal for Bacillus subtilis subsp. subtilis subsp. nov. and Bacillus subtilis subsp. spizizenii subsp. nov. Int J Syst Bacteriol 49:1211–1215. https://doi.org/10.1099/00207713-49-3-1211
Peypoux F, Bonmatin JM, Wallach J (1999) Recent trends in the biochemistry of surfactin. Appl Microbiol Biotechnol 51:553–563. https://doi.org/10.1007/s002530051432
Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T (2011) Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res 21(4):599–609. https://genome.cshlp.org/content/21/4/599
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Spieß T, Korn SM, Kötter P, Entian K-D (2015) Autoinduction specificities of the lantibiotics subtilin and nisin. Appl Environ Microbiol 81:7914–7923. https://doi.org/10.1128/AEM.02392-15
Stein T (2005) Bacillus subtilis antibiotics: structures, syntheses, and specific functions. Mol Microbiol 56:845–857. https://doi.org/10.1111/j.1365-2958.2005.04587.x
Stein T (2020) Oxygen-limiting growth conditions and deletion of the transition state regulator protein Abrb in Bacillus subtilis 6633 result in an increase in subtilosin production and a decrease in subtilin production. Probiotics Antimicrob Proteins 12:725–731. https://doi.org/10.1007/s12602-019-09547-4
Stein T, Entian K-D (2002) Maturation of the lantibiotic subtilin: matrix-assisted laser desorption/ionization time-of-flight mass spectrometry to monitor precursors and their proteolytic processing in crude bacterial cultures. Rapid Commun Mass Spectrom 16:103–110. https://doi.org/10.1002/rcm.552
Stein T, Borchert S, Kiesau P, Heinzmann S, Klöss S, Klein C, Helfrich M, Entian K-D (2002b) Dual control of subtilin biosynthesis and immunity in Bacillus subtilis. Mol Microbiol 44:403–416. https://doi.org/10.1046/j.1365-2958.2002.02869.x
Stein T, Düsterhus S, Stroh A, Entian K-D (2004) Subtilosin production by two Bacillus subtilis subspecies and variance of the sbo-alb cluster. Appl Environ Microbiol 70:2349–2353. https://doi.org/10.1128/AEM.70.4.2349-2353.2004
Stein T, Heinzmann S, Düsterhus S, Borchert S, Entian K-D (2005) Expression and functional analysis of the subtilin immunity genes spaIFEG in the subtilin-sensitive host Bacillus subtilis MO1099. J Bacteriol 187:822–828. https://doi.org/10.1128/JB.187.3.822-828.2005
Stein T (2008) Whole-cell matrix-assisted laser desorption/ionization mass spectrometry for rapid identification of bacteriocin/lantibiotic-producing bacteria. Rapid Commun Mass Spectrom 22:1146–1152. https://onlinelibrary.wiley.com/doi/full/10.1002/rcm.3481
Stein T, Borchert S, Conrad B, Feesche J, Hofemeister B, Hofemeister J, Entian K-D (2002a) Two different lantibiotic-like peptides originate from the ericin gene cluster of Bacillus subtilis A1/3. J Bacteriol 184:1703-1711. https://jb.asm.org/content/184/6/1703
Stein T, Heinzmann S, Kiesau P, Himmel B, Entian K-D (2003). The spa-box for transcriptional activation of subtilin biosynthesis and immunity in Bacillus subtilis. Mol Microbiol 47:1627–1636. https://onlinelibrary.wiley.com/doi/full/10.1046/j.1365-2958.2003.03374.x
Sun P, Li J, Bu D, Nan X, Du H (2016) Effects of Bacillus subtilis natto and different components in culture on rumen fermentation and rumen functional bacteria in vitro. Curr Microbiol 72:589–595. https://doi.org/10.1007/s00284-016-0986-z
Torres MJ, Brandan CP, Petroselli G, Erra-Balsells R, Audisio MC (2016) Antagonistic effects of Bacillus subtilis subsp. subtilis and B. amyloliquefaciens against Macrophomina phaseolina: SEM study of fungal changes and UV-MALDI-TOF MS analysis of their bioactive compounds. Microbiol Res 182:31–39. https://doi.org/10.1016/j.micres.2015.09.005
Tran C, Cock IE, Chen X, Feng Y (2022) Antimicrobial Bacillus: metabolites and their mode of action. Antibiotics 11:88. https://doi.org/10.3390/antibiotics11010088
Walker MC, Eslami SM, Hetrick KJ, Ackenhusen SE, Mitchell DA, van der Donk WA (2020) Precursor peptide-targeted mining of more than one hundred thousand genomes expands the lanthipeptide natural product family. BMC Genomics 21:387. https://doi.org/10.1186/s12864-020-06785-7
Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. https://doi.org/10.1093/bioinformatics/btp033
Yue X, Li LY, Zhang X, Zheng Y (2021) Microbial diversity and function of soybean paste in EastAsia: what we know and what we don’t. Curr Opin Food Sci 37:145–152. https://doi.org/10.1016/j.cofs.2020.10.012
Zeigler DR (2011) The genome sequence of Bacillus subtilis subsp. spizizenii W23: insights into speciation within the B. subtilis complex and into the history of B. subtilis genetics. Microbiology 157:2033–2041. https://doi.org/10.1099/mic.0.048520-0
Zeigler DR, Prágai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB (2008) The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol 190:6983–9695. https://jb.asm.org/content/190/21/6983
Zhang Q, Kobras CM, Gebhard S, Mascher T, Wolf D (2022) Regulation of heterologous subtilin production in Bacillus subtilis W168. Microb Cell Fact 21:57. https://doi.org/10.1186/s12934-022-01782-9
Zhang Q, Yu Y, Vélasquez JE, van der Donk WA (2012) Evolution of lanthipeptide synthetases. Proc Natl Acad Sci U S A. 109:18361–18366. https://www.pnas.org/content/109/45/18361
Zhao P, Xue Y, Gao W, Li J, Zu X, Fu D et al (2018) Bacillaceae-derived peptide antibiotics since 2000. Peptides 101:10–16. https://doi.org/10.1016/j.peptides.2017.12.018
Zheng G, Yan LZ, Vederas JC, Zuber P (1999) Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J Bacteriol 181:7346–7355. https://doi.org/10.1128/JB.181.23.7346-7355.1999
Acknowledgements
We greatly acknowledge Michael Karas, University Frankfurt, for the opportunity to use his MALDI-TOF mass spectrometric equipment.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
M.H. performed the research, interpreted the data, and wrote the paper; K.-D. E. conceived of or designed the study and reviewed the manuscript; and T. S. conceived of or designed the study and performed the research, interpreted the data, and wrote the paper. The authors read, reviewed, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent for publication
The authors declare that they have no conflict of interest.
Competing interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Helfrich, M., Entian, KD. & Stein, T. Antibiotic profiling of wild-type bacilli led to the discovery of new lanthipeptide subtilin-producing Bacillus spizizenii strains whose 16S rDNA sequences differ from the B. spizizenii typing strain. Int Microbiol 25, 839–850 (2022). https://doi.org/10.1007/s10123-022-00266-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-022-00266-5