Abstract
Background
Long non-coding RNAs (lncRNAs) are increasingly investigated in numerous carcinomas containing gastric cancer (GC). The aim of our research is to inquire about the expression profile and role of LBX2-AS1 in GC.
Methods
The expressions of LBX2-AS1, miR-219a-2-3p, FUS and LBX2 were measured by qRT-PCR. Western blot evaluated FUS and LBX2 protein levels. Cell proliferation and apoptosis were, respectively, evaluated by CCK-8, colony formation, EdU, flow cytometry and TUNEL assays. FISH and subcellular fractionation assays examined the position of LBX2-AS1. The binding between genes were certified by RIP, RNA pull-down, ChIP and luciferase reporter assays. Pearson correlation analysis analyzed the association of genes. Kaplan–Meier method detected the relationship of LBX2-AS1 expression with overall survival.
Results
The up-regulation of LBX2-AS1 in GC tissues and cells was verified. Function assays proved that LBX2-AS1 down-regulation restricted the proliferation ability. Then, we unveiled the LBX2-AS1/miR-219a-2-3p/FUS axis. Additionally, LBX2-AS1 positively regulated LBX2 mRNA stability via FUS. LBX2 transcriptionally modulated LBX2-AS1. In the end, rescue and in vivo experiments validated the whole regulatory mechanism.
Conclusion
LBX2-AS1/miR-219a-2-3p/FUS/LBX2 positive feedback loop mainly affected the proliferation and apoptosis abilities of GC cells, offering novel therapeutic targets for the treatment of patients with GC.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is a prevalent malignancy and also one of the leading causes of cancer-generated death all around the world [1,2,3]. Like other carcinomas, GC is featured by infinite cell proliferation and tumor growth, which is attributed to the promotive influences of oncogenic regulators and the inhibitory influences of tumor-suppressive regulators [4,5,6]. In the current time, the main treatment methods for GC patients are surgical operation, chemotherapy and radiotherapy [7, 8]. Considering the low diagnosis rate and high recurrence rate of GC, it is imperative to investigate more GC therapeutic targets to fight against the serious disease for human beings.
It is noted that dysregulation of non-coding RNAs (ncRNAs) exerts pivotal properties in human tumors, GC included. For example, lncRNA GHET1 overexpression predicts a dismal survival rate and clinical parameters of patients in different cancers [9]. LINC00441 binds to DNMT1 to modulate the methylation of RB1 in GC [10]. FER1L4/miR-372/E2F1 axis serves as a ceRNA model to affect the proliferation and cell cycle in glioma [11]. LncRNA UCA1 enhances tumor metastasis by targeting miR-203/ZEB2 pathway in gastric cancer [12]. In this paper, LBX2 antisense RNA 1 (LBX2-AS1), a newly reported carcinogenic lncRNA in esophageal squamous cell carcinoma, was probed [13]. The expression profiling and function role of LBX2-AS1 in GC has barely been explored till now.
Fused in sarcoma (FUS) has been shown to be oncogenic in numerous tumors, promoting cell proliferation and inhibiting cell apoptosis [14, 15]. For instance, NEAT1 is needed for the survival of breast cancer cells by miR-548/FUS pathway [16]. A cytoplasmic lncRNA LINC00470 acts as a AKT activator to promote glioblastoma cell autophagy [17]. LncRNA XIST facilitates cervical cancer progression through upregulation of FUS by competitive binding to miR-200a [18]. However, more detailed mechanisms around FUS involved in cancers are unclear.
The purpose of our research is to investigate the expression and molecular mechanism of LBX2-AS1 in GC. In consistent with the high expression of LBX2-AS1 in stomach adenocarcinoma (STAD) tissues, LBX2-AS1 was higher in GC tissues and cells. LBX2-AS1 silencing suppressed cell proliferation and exacerbated apoptosis. Based on the cytoplasm location of LBX2-AS1, we detected the LBX2-AS1/miR-219a-2-3p/FUS axis in GC. Additionally, LBX2-AS1 was found to positively regulate LBX2 via FUS. Meanwhile, LBX2 was proved to transcriptionally modulate LBX2-AS1. Eventually, rescue assay and in vivo experiments validated the whole regulatory mechanism.
Materials and methods
Tissue samples and cell lines
40 pairs of human GC tissues and adjacent normal tissues were acquired from GC patients who were diagnosed at the First Affiliated Hospital of Zhengzhou University. The experiment was permitted by the Research Medical Ethics Committee of the First Affiliated Hospital of Zhengzhou University and the written informed consent was signed by each GC patient. Prior to surgery, none of the patients was exposed to chemotherapy or radiotherapy. Harvested tissues were immediately snap-frozen at − 80 °C in liquid nitrogen and kept until RNA extraction. This work was carried out obeying the Ethical requirements of the Declaration of Helsinki. Human gastric mucosa epithelial cell (GES-1) and GC cells (MGC-803, BGC-823, SGC-7901, AGS) were obtained from the Chinese Academy of Sciences (Shanghai, China) and cultivated in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) with 10% FBS (Gemini Bioproducts, Calabasas, CA) in a moist condition with 5% CO2 at 37 °C.
Cell transfection
AGS and SGC-7901 cells were plated into 6-well plates and grown to 80% confluence. Cells were transfected with shRNAs against LBX2-AS1 (shLBX2-AS1#1/2/3), FUS (shFUS#1/2/3) and LBX2 (shLBX2) and negative controls (shNCs) for RNA interference. shRNAs’ sequences were shown as follows: shNC: CCGCCTTAATGTGCAATAAAGCAGCCTCGAGGCTGCTTTATTGCACATTAAGTTTTTG; shLBX2-AS1#1: CCGCGGAATGTTTGCTGAATTAATGCTCGAGCATTAATTCAGCAAACATTCCTTTTTG; shLBX2-AS1#2: CCGCCCATTTGTTTTTTAGACTTCTCTCGAGAGAAGTCTAAAAAACAAATGGTTTTTG; shLBX2-AS1#3: CCGCCAAGTTATAAAACTATAATGCCTCGAGGCATTATAGTTTTATAACTTGTTTTTG; shFUS#1: CCGCGATAATTCAGACAACAACACCCTCGAGGGTGTTGTTGTCTGAATTATCTTTTTG; shFUS#2: CCGCGCTAAAGCAGCTATTGACTGGCTCGAGCCAGTCAATAGCTGCTTTAGCTTTTTG; shFUS#3: CCGCCTATGTAATTGTAACTATACCCTCGAGGGTATAGTTACAATTACATAGTTTTTG; shLBX2: CCGCGTCTTCAATCGTCCACCTGTACTCGAGTACAGGTGGACGATTGAAGACTTTTTG. The pcDNA3.1 vector expressing LBX2 or FUS and the empty pcDNA3.1 vector, as well as miR-219a-2-3p mimics and NC mimics were utilized for RNA overexpression. All above plasmids were bought from Genechem (Shanghai, China). Cells were reaped at 48 h after transfection using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Quantitative real-time PCR (qRT-PCR)
qRT-PCR was conducted as before [19]. GAPDH and U6 acted as the endogenous normalizations. PCR primers were listed as follows: LBX2-AS1 (Forward): AGTTTGTCCCAGGTTTGGCA; LBX2-AS1 (Reverse): CATGCCAGGGTCCTTGTTCT; FUS (Forward): CTATGGACAGCAGGACCGTG; FUS (Reverse): CCTCTGGGTTCATAGCCACC; miR-219a-2-3p (Forward): AGAAUUGUGGCUGGACAUCUGU; miR-219a-2-3p (Reverse): CTCTACAGCTATATTGCCAGCCAC; LBX2 (Forward): TAAGCTGCACAGGACTTCCG; LBX2 (Reverse): CAGGTGGTCACTTGGTTCCA; GAPDH (Forward): CCACATCGCTCAGACACCAT; GAPDH (Reverse): TGACAAGCTTCCCGTTCTCA; U6 (Forward): CTCGCTTCGGCAGCACA; U6 (Reverse): AACGCTTCACGAATTTGCGT.
Bioinformatics analysis
Through browsing GEPIA (http://gepia.cancer-pku.cn/index.html), the relatively high expression of LBX2-AS1 in multiple malignancies, including stomach adenocarcinoma (STAD) was observed and identified. MiRNAs that had complementary binding sites with LBX2-AS1 were predicted from starBase v(http://starbase.sysu.edu.cn/). Downstream target mRNAs of miR-219a-2-3p were also obtained from starBase. DNA motif of LBX2 and its binding sites with LBX2-AS1 promoter were downloaded from JASPAR (http://jaspar.genereg.net/). The cytoplasmic or nuclear location of LBX2-AS1 was analyzed using lnclocator (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/). Genomic position of LBX2 or LBX2-AS1 was identified by screening UCSC genome browser (http://genome.ucsc.edu/).
Actinomycin D assay
Transfected cells were treated with Actinomycin D (Sigma-Aldrich) and analyzed by qRT-PCR at specific time points.
Cell proliferation assays
For colony formation, cells were seeded into 6-well plates. 2 weeks later, colonies were fixed in methanol (Solarbio, Beijing, China) and dyed by crystal violet (Solarbio), followed by manual calculation. CCK-8 and EdU staining were performed as described previously [20].
Cell apoptosis assays
AGS and SGC-7901 cell apoptosis was evaluated using Click-iT® TUNEL Alexa Fluor® Imageing Assay (Invitrogen). Cells were stained with DAPI (Thermo Fisher Scientific) and observed by Olympus IX70 Inverted Microscope (Olympus, Tokyo, Japan). For flow cytometry analysis, cells were subjected to FITC Annexin V Apoptosis Detection Kit (Ruibo, Guangzhou, China) for 30 min in the dark, evaluated by BD Accuri C6 Plus personal flow cytometer (Becton–Dickinson, Mountain View, USA).
Fluorescence in situ hybridization (FISH) assay
LBX2-AS1 subcellular localization was evaluated by FISH Tag™ DNA Multicolor Kit, Alexa Fluor™ dye combination (Thermo fisher, F32951). Cells were fixed by 4% paraformaldehyde. LBX2-AS1 FISH probe (Sigma-Aldrich, Milan, Italy) hybridization solution was added into plate. Nucleus was stained with DAPI for 10 min. Laser Confocal Laser Scanning Microscope-FV3000 (Olympus) was used to obtain the fluorescence images.
Subcellular fractionation assay
Cytoplasmic and Nuclear RNA Purification Kit (Norgen, Ontario, Canada) was employed based on the specification. Expression levels of LBX2-AS1 were assessed by qRT-PCR analysis. GAPDH and U6 were used for cytoplasmic or nuclear control.
RNA immunoprecipitation (RIP) and RNA pull-down assays
MS2-RIP and Ago2-RIP were carried out based on the previous study [21]. As for RNA pull-down assay, cell lysates were incubated with biotinylated RNA overnight, then cultivated with streptavidin agarose beads. Finally, the pulled-down complex was assayed by qRT-PCR or western blotting.
Luciferase reporter assay
The wild-type or mutant binding sites of miR-219a-2-3p in LBX2-AS1 sequences or FUS 3′ UTR were sub-cloned into pmirGLO dual-luciferase vector, named as LBX2-AS1-WT/MUT or FUS-WT/MUT, followed by co-transfection with miR-219a-2-3p mimics or NC mimics into AGS or SGC-7901 cells. The pGL3-Basis luciferase vector (Promega) was utilized for the LBX2-AS1 promoter-luciferase analysis. The luciferase activity was measured after 48 h using Dual-Luciferase Report Assay System (Promega, Madison, WI, USA).
ChIP assay
Magna ChIP Kit (Millipore) was purchased for ChIP assay. After cross-linked chromatin fragments were sonicated, lysates were immunoprecipitated with anti-LBX2 or anti-IgG antibody. Precipitated chromatin was detected by qRT-PCR. Primers for ChIP-qPCR were as follows: Forward: TTCTACTCTCCCCTGAGCCC; Reverse: ACTCCCCTTCCTGCGAATTG.
In vivo tumor growth assay
Six-week-old male BALB/C athymic nude mice were commercially acquired from National Laboratory Animal Center (Beijing, China). Animal experiment was conducted strictly in light of the protocol approved by the Administrative Panel on Laboratory Animal Care of the First Affiliated Hospital of Zhengzhou University. Ethical approval has been provided by the institute. The AGS cells transfected with shLBX2-AS1#1 or shNC were collected and subcutaneously injected into nude mice, with the measurement of tumor volume every 4 days. 28 days after injection, mice were killed. Tumors were excised and weighed for subsequent analysis.
Immunohistochemistry staining (IHC)
Tumor tissue samples from in vivo assay were fixed by 4% paraformaldehyde and immunostained with Hematoxylin & Eosin (H&E). IHC staining was conducted on consecutive 4 μm thick sections of paraffin-embedded tissues. The specific antibody against Ki67 (ab15580, abcam), PCNA (ab92552, abcam), FUS (ab124923, abcam) or LBX2 (PA5-60662, ThermoFisher) was obtained.
Western blot
Cells were lysed by RIPA buffer, subjected to SDS-PAGE and transferred to PVDF (Millipore, MA, USA). After sealing with skim milk, membranes were incubated with primary antibodies including anti-FUS (ab228840, Abcam, Cambridge, USA), anti-LBX2 (ab164764, Abcam) and anti-GAPDH (ab181602, Abcam). Proteins were detected using chemiluminescence system.
Statistical analysis
Data from three replications were analyzed by SPSS 22.0 (SPSS, Chicago, USA) and GraphPad Prism 6 (GraphPad, San Diego, CA) and denoted as mean ± SD. Overall survival was calculated by Kaplan–Meier method. Gene expression correlation was assayed by Pearson’s correlation analysis. Significance of variances was evaluated by Student’s t test or ANOVA. P < 0.05 has statistical significance.
Results
LBX2-AS1 was overexpressed in gastric cancer and its knockdown restrained cell proliferation and stimulated apoptosis
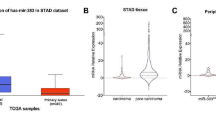
Through browsing GEPIA dataset, we observed the relatively high expression of LBX2-AS1 in multiple malignancies, including stomach adenocarcinoma (STAD) (Supplementary Fig. 1a, b and Fig. 1a). Our study aimed to explore possible molecular targets for gastric cancer (GC), hence we chose LBX2-AS1 for in-depth investigations. Accordingly, LBX2-AS1 expression was significantly elevated in GC tissues in comparison with normal tissues (Fig. 1b). Kaplan–Meier analysis discovered that high LBX2-AS1 expression was in association with low survival rate (Fig. 1c). Similarly, LBX2-AS1 expression was also distinctly heightened in GC cells compared to normal GES-1 cells (Fig. 1d). Sequentially, loss-of-function assays were conducted for determining the influences of LBX2-AS1 on GC cells. The shRNAs targeting LBX2-AS1 (shLBX2-AS1#1/2/3) were transfected into AGS and SGC-7901 cells with higher levels of LBX2-AS1 to knock down LBX2-AS1 expression (Fig. 1e). CCK-8 assay demonstrated that cell viability was inhibited when LBX2-AS1 was silenced (Fig. 1f). Colony forming and EdU assays found that cell proliferation was repressed after silencing LBX2-AS1 (Fig. 1g, h). Via flow cytometry analysis, it was viewed that cell apoptosis rate was raised by LBX2-AS1 depletion, which was in accordant with the results of TUNEL assay (Fig. 1i, j). Totally, cell proliferation and apoptosis were, respectively, suppressed and promoted by LBX2-AS1 inhibition in GC.
LBX2-AS1 depletion negatively regulated GC cell proliferation and positively modulated apoptosis. a, b LBX2-AS1 was upregulated in GC tissue samples from TCGA dataset or resected GC tumor tissues. c Kaplan–Meier analysis showed that high LBX2-AS1 expression predicted poor prognosis of GC patients. d Upregulation of LBX2-AS1 in GC cells. e LBX2-AS1 expression was weakened with the specific shRNAs transfection in AGS and SGC-7901 cells. f, h CCK-8, colony formation and EdU assays presented the inhibition of shLBX2-AS1#1 on proliferative ability of AGS and SGC-7901 cells. i, j Flow cytometry and TUNEL assay presented that LBX2-AS1 knockdown induced more apoptotic GC cells. Results were expressed as the mean ± SD. *P < 0.05, **P < 0.01
LBX2-AS1 exerted as a sponge for miR-219a-3p in GC
The function roles of lncRNAs are largely due to their locations in tumor cells [22, 23]. And LBX2-AS1 was assumed to be located in the cytosol of GC cells with the assistance of lncLocator website (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/) (Supplementary Fig. 1c). Then FISH and subcellular fractionation experiments were employed to locate LBX2-AS1 in AGS and SGC-7901 cells. The results indicated that LBX2-AS1 was mainly seated in the cytoplasm of GC cells (Fig. 2a, b). The ceRNA network is a well-known model constituted by lncRNAs/circRNAs, microRNAs and mRNAs in the cytoplasm [24, 25], which prompted us to inquire about the associated mechanism. RIP experiment detected that a large proportion of LBX2-AS1 was precipitated by anti-Ago2, hinting the participation of LBX2-AS1 in ceRNA model (Fig. 2c). Therefore, we utilized starBase, an online prediction tool for RNA interactions, to look for downstream miRNAs. From starBase, 23 predicted miRNAs binding with LBX2-AS1 were found out. We analyzed the levels of these miRNAs in tumor versus control cells. Results showed that seven miRNAs (miR-4784, miR-3150a-3p, miR-384, miR-665, miR-650, miR-491-5p and miR-219a-2-3p) were overtly down-regulated (Supplementary Table 1). Next, MS2-pull-down assay was applied to examine which miRNA possessed the highest binding affinity with LBX2-AS1. Consequently, miR-219a-2-3p represented the highest enrichment by MS2-LBX2-AS1-wt probe (Fig. 2d). For luciferase reporter assay, the binding sites between LBX2-AS1 and miR-219a-2-3p were shown and miR-219a-2-3p was up-regulated by miR-219a-2-3p mimics (Fig. 2e, f). The luciferase activity of LBX2-AS1-WT was refrained by miR-219a-2-3p overexpression and that of LBX2-AS1-MUT varied not (Fig. 2g). Additionally, miR-219a-2-3p was markedly repressed in GC tissues, and Pearson correlation analysis revealed that miR-219a-2-3p expression was negatively associated with LBX2-AS1 expression (Fig. 2h, i). In GC cells, miR-219a-2-3p levels were dramatically down-regulated (Fig. 2j). What’s more, LBX2-AS1 expression was silenced with miR-219a-2-3p overexpression and miR-219a-2-3p expression was increased with LBX2-AS1 down-regulation (Fig. 2k, l). These findings illustrated that miR-219a-2-3p was–negatively regulated by LBX2-AS1.
LBX2-AS1 sponged miR-219a-2-3p in GC cells. a, b FISH and subcellular location assay indicated that LBX2-AS1 was mainly enriched in the cytoplasm of AGS and SGC-7901 cells. c LBX2-AS1 abundance in precipitated Ago2 complex was assessed by RIP assay. d MS2-pull-down assay revealed the miRNAs interacting with LBX2-AS1 in GC cells. e Luciferase reporters were established based on predicted binding sites between miR-219a-2-3p and LBX2-AS1. f Transfection efficiency of miR-219a-2-3p mimics. g Luciferase reporter assay demonstrated the binding miR-219a-2-3p and LBX2-AS1-WT. h qRT-PCR analysis proved the downregulation of miR-219a-2-3p in GC tissue samples. i The negative expression correlation of miR-219a-2-3p with LBX2-AS1. j qRT-PCR analysis proved the downregulation of miR-219a-2-3p in GC cells. k qRT-PCR analysis probed the LBX2-AS1 expression in the presence of miR-219a-2-3p mimics. l qRT-PCR analysis examined miR-219a-2-3p expression in response to shLBX2-AS1#1. Results were expressed as the mean ± SD. *P < 0.05, **P < 0.01
miR-219a-2-3p targeted FUS which affected the proliferation and apoptosis of GC cells
MicroRNAs have been widely reported as tumor suppressors by regulating their downstream mRNAs [26, 27]. In this s regard, we analyzed the downstream targets of miR-219a-3p. Then top 200 target mRNAs (AgoExpNum) of miR-219a-2-3p were unveiled from starBase v2.0 (http://starbase.sysu.edu.cn/). The levels of 200 mRNAs were compared in GES-1, AGS, and SGC-7901 cells by qRT-PCR analysis to obtain the differentially expressed genes. The top five up-regulated mRNAs were FUS, SHOC2, FNDC3B, G3BP2 and NUFIP2 (Fig. 3a). Subsequently, we examined five gene expressions in GC tissues. FUS was highly expressed in GC tissues (Supplementary Fig. 1d–g and Fig. 3b). Pearson correlation analysis determined that FUS was positively correlated with LBX2-AS1 but inversely associated with miR-219a-2-3p (Fig. 3c). FUS mRNA and protein levels were also higher in GC cells than that in GES-1 cells (Fig. 3d). In addition, the mRNA and protein levels of FUS were lowered by LBX2-AS1 knockdown or miR-219a-2-3p overexpression (Fig. 3e, f). After confirming the impact of LBX2-AS1 or miR-219a-2-3p on FUS expression, we attempted to verify the binding between miR-219a-2-3p and FUS mRNA. In RNA pull-down experiment, FUS was pulled down by bio-miR-219a-2-3p-wt (Fig. 3g). Bioinformatics tools predicted the interaction between miR-219a-2-3p and FUS mRNA (Fig. 3h). Luciferase reporter assay presented miR-219a-2-3p mimics impaired the luciferase activity of FUS-WT (Fig. 3i). Besides, the functional role of FUS in GC cells was probed. FUS expression at mRNA and protein levels was, respectively, knocked down by shFUS#1, shFUS#2 and shFUS#3, among which shFUS#2 caused the lowest FUS levels (Fig. 3j). The results of CCK-8 and EdU assays indicated that FUS silencing suppressed cell proliferation (Fig. 3k, l). Flow cytometry analysis showed that cell apoptosis was activated upon FUS knockdown (Fig. 3m). These data elucidated that FUS was targeted by miR-219a-2-3p and upregulated by LBX2-AS1.
FUS acted as the target gene of miR-219a-2-3p. a qRT-PCR analysis for the possible mRNAs influenced by miR-219a-2-3p. b qRT-PCR analysis verified the high expression of FUS in GC tissues. c Pearson’s correlation analysis presented the correlation between FUS and LBX2-AS1 or miR-219a-2-3p expressions. d qRT-PCR and western blot analyses showed the elevated mRNA and protein levels of FUS in GC cells. e, f qRT-PCR and western blot analyses disclosed the impacts of depleted LBX2-AS1 or enhanced miR-219a-2-3p on FUS expression AGS and SGC-7901 cells. g–i The combination of miR-219a-2-3p and FUS mRNA was verified by RNA pull-down and luciferase reporter assays. j qRT-PCR and western blot analyses of FUS expression after transfection with shRNAs against FUS in GC cells. k–m Cell viability, proliferation and apoptosis of AGS and SGC-7901 cells were monitored. Results were expressed as the mean ± SD. **P < 0.01
LBX2-AS1 positively regulated LBX2 via FUS
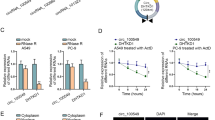
LBX2 was discovered to be a nearby gene of LBX2-AS1 by UCSC genome browser. Whether LBX2-AS1 could modulate LBX2 was not clear, thereafter we tested the levels of LBX2 under transfection of shLBX2-AS1#1. It was noted that LBX2 mRNA and protein levels were decreased following LBX2-AS1 deficiency (Fig. 4a). Then we performed RIP assay to observe the participation of LBX2-AS1 and LBX2 in ceRNA model. LBX2-AS1 and LBX2 were not co-expressed in Ago2 protein, indicating that LBX2-AS1 could not function as ceRNA to modulate LBX2 expression (Fig. 4b). Considering that RNA-binding protein (RBP) interaction is also a noted regulation mechanism in the cytoplasm [28]. We supposed that RBP may involve in LBX2-AS1-mediated LBX2 regulation. After the addition of Actinomycin D repressing RNA synthesis, the half-life of LBX2 was advanced by LBX2-AS1 down-regulation (Fig. 4c). To find the possible proteins binding with LBX2 mRNA, Pull-down silver staining and mass-spectrometry analysis were implemented. Strikingly, FUS protein was emerged among those proteins. Additionally, the binding of FUS protein to LBX2 was further confirmed by performing RNA pull-down assay followed by western blot analysis (Fig. 4d). Intriguingly, FUS silencing promoted the degradation of LBX2 mRNA under Actinomycin D addition (Fig. 4e). As shown in Fig. 4f, three assumed binding sites (LBX2-201, LBX2-202 and LBX2-203) of LBX2 mRNA for binding FUS were obtained. In RNA pull-down assay, FUS protein was only pulled down in LBX2-201 group (Fig. 4g). The binding of FUS to LBX2 was testified again by RIP assay (Fig. 4h). To observe the co-impacts of LBX2-AS1 and FUS on LBX2, FUS was overexpressed by pcDNA3.1/FUS (Fig. 4i). As a result, LBX2 mRNA stability was reduced by shLBX2-AS1#1 but regained by pcDNA3.1/FUS (Fig. 4j). Above results testified the hypothesis told us that LBX2-AS1 positively modulated LBX2 via FUS.
LBX2-AS1 modulated LBX2 stability via FUS. a The mRNA and protein levels of LBX2 were diminished through transfecting cells with shLBX2-AS1#1. b LBX2-AS1 and LBX2 mRNA did not co-existed in Ago2 protein c LBX2 mRNA stability was destroyed by shLBX2-AS1#1 under Actinomycin D treatment. d Detection of the proteins binding to LBX2 by silver staining and RNA pull-down assays. e LBX2 mRNA stability was decreased by shFUS after treating cells with Actinomycin D. f Bioinformatics analysis predicted the binding region of LBX2 for FUS. g, h RNA pull-down and RIP assay proved the binding of FUS to LBX2 (3′UTR). i FUS overexpression vector strengthened the mRNA and protein levels of FUS in qRT-PCR analysis. j The reduced LBX2 mRNA stability by silencing LBX2-AS1 was restored via enhancing FUS expression in qRT-PCR assay. Results were expressed as the mean ± SD. *P < 0.05, **P < 0.01
LBX2 transcriptionally regulated LBX2-AS1
LBX2 has been reported to function as a transcription factor in cellular processes [29]. Interestingly, it was displayed that LBX2 knockdown or overexpression could negatively or positively mediate LBX2-AS1 (Fig. 5a, b). Further, we sought to explore whether LBX2 could serve as a transcription factor of LBX2-AS1. Herein, JASPAR was utilized to investigate the DNA motif of transcription factor LBX2. The DNA motif of LBX2 was exhibited in Fig. 5c. The wild and mutated binding sites of LBX2-AS1 promoter were presented in Fig. 5d. The luciferase activity of LBX2-AS1-WT (wild LBX2-AS1 promoter) or LBX2-AS1-Mut2 (LBX2-AS1 promoter with mutated P2) was significantly injured by the knockdown of LBX2, whereas that of LBX2-AS1-Mut1 (LBX2-AS1 promoter with mutated P1) or LBX2-AS1-Mut1 + 2 (LBX2-AS1 promoter with mutated P1 and P2) changed not, indicating P1 was the actual binding sites (Fig. 5e). ChIP assay validated that LBX2 was largely enriched LBX2-AS1 promoter (Fig. 5f). We then estimated the levels of LBX2 in GC tissues. LBX2 expression was obviously up-regulated in GC tissues (Fig. 5g). Additionally, LBX2 expression and LBX2-AS1 expression were in positive correlation (Fig. 5h). Moreover, LBX2 mRNA and protein levels were also heightened in GC cells (Fig. 5i). These conclusions hinted that LBX2-AS1 was under the transcriptional regulation of LBX2.
LBX2 affected LBX2-AS1 transcription in GC cells. a AGS and SGC-7901 cells transfected with shLBX2 or pcDNA3.1/LBX2 were subjected to western blotting. b The expression changes of LBX2-AS1 in response to shLBX2 or pcDNA3.1/LBX2. c The DNA motif of LBX2 was shown. d The binding sequences between LBX2 protein and LBX2-AS1 promoter were shown. e, f Luciferase reporter and ChIP assays identified LBX2 as the transcription factor of LBX2-AS1. g qRT-PCR showed that LBX2 was expressed highly in GC tissues. h The positive correlation between LBX2-AS1 and LBX2was revealed by Pearson’s correlation analysis. i The mRNA and protein levels of LBX2 in GC cells. Results were expressed as the mean ± SD. *P < 0.05, **P < 0.01
FUS or LBX2 overexpression abrogated the inhibitory effect of LBX2-AS1 silence on the proliferation of GC cells
In the end, rescue assays were carried out to confirm the whole modulatory mechanism. In CCK-8 assay, cell viability inhibited by shLBX2-AS1#1 was regained by pcDNA3.1/FUS or pcDNA3.1/LBX2 (Fig. 6a). In colony forming assay, the number of colonies was decreased upon LBX2-AS1down-regulation but increased via FUS or LBX2 promotion (Fig. 6b). EdU assay exhibited that LBX2-AS1 silence repressed cell proliferation and this phenomenon was neutralized by overexpression of FUS or LBX2 (Fig. 6c). According to flow cytometry analysis, the increased cell apoptosis by LBX2-AS1 silencing was reduced by FUS or LBX2 enhancement (Fig. 6d). TUNEL assay indicated that the number of TUNEL positive cells was increased by LBX2-AS1 knockdown which was abolished with the introduction of FUS or LBX2 (Fig. 6e). All these experiments proved that LBX2-AS1 regulated GC proliferation via FUS and LBX2.
LBX2-AS1 contributed to GC cell proliferation through miR-219a-2-3p/FUS/LBX2 axis. a–c Cell proliferation assays implied that the proliferative capacity of AGS cells was weakened by LBX2-AS1 knockdown, but was rescued after overexpressing FUS or LBX2. d, e Flow cytometry and TUNEL manifested that overexpression of FUS or LBX2 could decrease the amount of LBX2-AS1 depletion-mediated apoptotic cells. Results were expressed as the mean ± SD. **P < 0.01
LBX2-AS1 promoted tumor growth of GC in vivo
To explore whether LBX2-AS1 could affect GC growth in vivo, we established animal models via the injection of AGS cells pre-treated with shNC or shLBX2-AS1#1 into mice. The volume of xenograft tumors was measured every four days. 28 days later, tumor weight was also weighed. As presented in Fig. 7a, shLBX2-AS1#1 group tumor developed slower than that in sh-NC group. Tumor weight in shLBX2-AS1#1 group was lighter compared with shNC group (Fig. 7b). qRT-PCR in xenograft tumors demonstrated that LBX2-AS1, FUS and LBX2 levels were reduced in shLBX2-AS1#1 group while the level of miR-219a-2-3p was augmented (Fig. 7c). Western blot assay also proved the decreased protein level of FUS and LBX2 in shLBX2-AS1#1 group (Fig. 7d). IHC staining discovered that Ki67 and PCNA positivity was lowered in LBX2-AS1 knockdown tumors. FUS and LBX2 expression was accordingly declined in LBX2-AS1 knockdown tumors (Fig. 7e). In conclusion, LBX2-AS1 stimulated tumor growth of GC in vivo.
LBX2-AS1 promoted GC cell growth in vivo. a Tumor growth curves of mice in two groups. b Tumor weights in two groups. c qRT-PCR assay of LBX2-AS1, miR-219a-2-3p, FUS, and LBX2 level in mouse tumors from two groups. d Western blot assay of FUS and LBX2 protein in mouse tumors from two groups. e (h & e) staining and immunohistochemical (IHC) assay detection of Ki67, PCNA, FUS and LBX2 expressions. f Schematic model of the LBX2-AS1/miR-219a-2-3p/FUS/LBX2 positive feedback loop in gastric cancer. Results were expressed as the mean ± SD. *P < 0.05, **P < 0.01
Discussion
The explorations of long non-coding RNAs (lncRNAs) in gastric cancer (GC) are broadly conducted. For example, LINC00707 promotes proliferation and metastasis in gastric cancer via interaction with HuR [30]. The lncRNA HOTAIR/miR-17-5p/PTEN axis functions inversely with Shenqifuzheng (SQFZ) injection in relation to the chemosensitivity of GC cells [31]. LINC01234 sponges miR-204-5p to regulate CBFB expression by acting as a competing endogenous RNA in gastric cancer [32]. Nevertheless, more studies of functional lncRNAs are requisite.
Previous documentation revealed that elevated LBX2-AS1 facilitates esophageal squamous cell carcinoma cell migration and EMT [13]. Besides, LBX2-AS1 was increased in non-small cell lung cancer and correlated with poor prognosis and malignant cellular behaviors [33]. Via browsing the cancer genome atlas (TCGA) dataset, we viewed that LBX2-AS1 antisense RNA 1 (LBX2-AS1) was an overexpressed lncRNA in nearly 70% of cancers, implying its huge potential in tumor regulation. In stomach adenocarcinoma (STAD), LBX2-AS1 expression was also heightened. Currently, our group has concentrated on gastric cancer development. Taken these data together, we determined to explore the undefined expression pattern and possible role of LBX2-AS1 in GC. For the first time, our study revealed the LBX2-AS1 was highly expressed and played oncogenic function in GC. LBX2-AS1 depletion attenuated the unlimited cell growth, migration and EMT process while increased cell apoptosis in GC.
Subsequently, we decided to excavate the specified mechanism underlying LBX2-AS1. Due to the cytoplasm location of LBX2-AS1, ceRNA model occurred in our mind where lncRNAs can serve as sponges of microRNAs (miRNAs) targeting downstream genes. For instances, LINC00665 boosts the progression of lung adenocarcinoma by regulating AKR1B10-ERK signaling through sponging miR-98 [34]; lncRNA NORAD activate the TGF-β pathway to facilitate hepatocellular carcinoma progression by miR-202-5p [35]; STAT3-induced upregulation of lncRNA MEG3 regulates the growth of cardiac hypertrophy through miR-361-5p/HDAC9 axis [36]. Hence, 23 predicted target miRNAs of LBX2-AS1 obtained from starBase were analyzed by qRT-PCR. Seven miRNAs were selected and further experimented to find the miRNA binding with LBX2-AS1 to the largest extent. MS2-pull-down assay determined that miR-219a-2-3p owned the highest affinity with LBX2-AS1. Mechanism experiments affirmed that miR-219a-2-3p was expressed at low levels in GC tissues and cells, and also in negative relation with LBX2-AS1. Further, we sought out fused in sarcoma (FUS), the most possible downstream gene under LBX2-AS1/miR-219a-2-3p axis. FUS has already been reported to promote tumor progression by various researches [16,17,18]. In this paper, FUS was highly expressed in GC tissues and cells. FUS expression was in positive association with LBX2-AS1 and in negative association with miR-219a-2-3p. In addition, silencing of FUS could repress cell proliferation and activate cell apoptosis. Here, we discovered the LBX2-AS1/miR-219a-2-3p/FUS pathway.
from UCSC website, we found that LBX2 was the host gene of LBX2-AS1. LBX2 was positively regulated by LBX2-AS1. Nevertheless, LBX2 and LBX2-AS1 was not as enriched in Ago2 protein. Eventually, we uncovered that the mRNA stability of LBX2 could be impaired when LBX2-AS1 was silenced. Since that, we supposed that there might be some RNA-binding protein (RBP) serving between LBX2-AS1 and LBX2. Strikingly, FUS was revealed to bind with LBX2. Then, LBX2-AS1 was proved to mediate LBX2 stability via FUS. What’s more, LBX2 has been demonstrated to exert its function as a transcription factor [29]. Thereafter, we determined that LBX2 could modulate LBX2-AS1 at transcriptional level. Overall, LBX2-AS1 post-transcriptionally modulated LBX2 via FUS and LBX2 in turn transcriptionally regulated LBX2-AS1.
Both rescue experiments and in vivo assays testified that LBX2-AS1/miR-219a-2-3p/FUS/LBX2 axis positively worked in GC growth. In conclusion, LBX2-AS1 absorbed miR-219a-2-3p to increase FUS, therefore, promoting FUS protein levels to stabilize LBX2 that could activate LBX2-AS1 (Fig. 7f). These findings provided us with new insights into the therapy of GC patients.
References
Liu L, Tian YC, Mao G, Zhang YG, Han L. MiR-675 is frequently overexpressed in gastric cancer and enhances cell proliferation and invasion via targeting a potent anti-tumor gene PITX1. Cell Signal. 2019;62:109352. https://doi.org/10.1016/j.cellsig.2019.109352(Epub 2019/07/02).
Hevia MJ, Castro P, Pinto K, Reyna-Jeldes M, Rodríguez-Tirado F, Robles-Planells C, et al. Differential effects of purinergic signaling in gastric cancer-derived cells through P2Y and P2X receptors. Front Pharmacol. 2019;10:612. https://doi.org/10.3389/fphar.2019.00612(Epub 2019/06/29).
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018. https://doi.org/10.3322/caac.21492(Epub 2018/09/13).
Li H, Wang Y. Long noncoding RNA (lncRNA) MIR22HG suppresses gastric cancer progression through attenuating NOTCH2 signaling. Med Sci Monit. 2019;25:656–65. https://doi.org/10.12659/msm.912813(Epub 2019/01/24).
Wei GH, Wang X. lncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(17):3850–6 (Epub 2017/10/05).
Li C, Liang G, Yang S, Sui J, Wu W, Xu S, et al. LncRNA-LOC101928316 contributes to gastric cancer progression through regulating PI3 K-Akt-mTOR signaling pathway. Cancer Med. 2019. https://doi.org/10.1002/cam4.2165(Epub 2019/06/18).
Kim BH, Eom KY, Kim JS, Kim HH, Park DJ. Role of salvage radiotherapy for regional lymph node recurrence after radical surgery in advanced gastric cancer. Radiat Oncol J. 2013;31(3):147–54. https://doi.org/10.3857/roj.2013.31.3.147(Epub 2013/10/19).
Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39(7):1010428317714626. https://doi.org/10.1177/1010428317714626(Epub 2017/07/04).
Jiang YF, Zhang HY, Ke J, Shen H, Ou HB, Liu Y. Overexpression of LncRNA GHET1 predicts an unfavourable survival and clinical parameters of patients in various cancers. J Cell Mol Med. 2019. https://doi.org/10.1111/jcmm.14486(Epub 2019/06/29).
Zhou J, Shi J, Fu X, Mao B, Wang W, Li W, et al. Linc00441 interacts with DNMT1 to regulate RB1 gene methylation and expression in gastric cancer. Oncotarget. 2018;9(101):37471–9. https://doi.org/10.18632/oncotarget.23928(Epub 2019/01/26)).
Xia L, Nie D, Wang G, Sun C, Chen G. FER1L4/miR-372/E2F1 works as a ceRNA system to regulate the proliferation and cell cycle of glioma cells. J Cell Mol Med. 2019;9:9–10. https://doi.org/10.1111/jcmm.14198(Epub 2019/03/20).
Gong P, Qiao F, Wu H, Cui H, Li Y, Zheng Y, et al. LncRNA UCA1 promotes tumor metastasis by inducing miR-203/ZEB2 axis in gastric cancer. Cell Death Dis. 2018;9(12):1158. https://doi.org/10.1038/s41419-018-1170-0(Epub 2018/11/23).
Zhang Y, Chen W, Pan T, Wang H, Zhang Y, Li C. LBX2-AS1 is activated by ZEB1 and promotes the development of esophageal squamous cell carcinoma by interacting with HNRNPC to enhance the stability of ZEB1 and ZEB2 mRNAs. Biochem Biophys Res Commun. 2019;511(3):566–72. https://doi.org/10.1016/j.bbrc.2019.02.079(Epub 2019/03/03).
Zhang Y, Huang Z, Sheng F, Yin Z. MYC upregulated LINC00319 promotes human acute myeloid leukemia (AML) cells growth through stabilizing SIRT6. Biochem Biophys Res Commun. 2019;509(1):314–21. https://doi.org/10.1016/j.bbrc.2018.12.133(Epub 2018/12/28).
Ward CL, Boggio KJ, Johnson BN, Boyd JB, Douthwright S, Shaffer SA, et al. A loss of FUS/TLS function leads to impaired cellular proliferation. Cell Death Dis. 2014;5(12):e1572. https://doi.org/10.1038/cddis.2014.508(Epub 2014/12/17).
Ke H, Zhao L, Feng X, Xu H, Zou L, Yang Q, et al. NEAT1 is required for survival of breast cancer cells through FUS and miR-548. Gene Regul Syst Bio. 2016;10(Suppl 1):11–7. https://doi.org/10.4137/grsb.s29414(Epub 2016/05/06).
Liu C, Zhang Y, She X, Fan L, Li P, Feng J, et al. A cytoplasmic long noncoding RNA LINC00470 as a new AKT activator to mediate glioblastoma cell autophagy. J Hematol Oncol. 2018;11(1):77. https://doi.org/10.1186/s13045-018-0619-z(Epub 2018/06/06).
Zhu H, Zheng T, Yu J, Zhou L, Wang L. LncRNA XIST accelerates cervical cancer progression via upregulating Fus through competitively binding with miR-200a. Biomed Pharmacother. 2018;105:789–97. https://doi.org/10.1016/j.biopha.2018.05.053(Epub 2018/06/18).
Ji Q, Li Y, Zhao Q, Fan LQ, Tan BB, Zhang ZD, et al. KLF11 promotes gastric cancer invasion and migration by increasing Twist1 expression. Neoplasma. 2019;66(1):92–100. https://doi.org/10.4149/neo_2018_180325n201(Epub 2018/12/05).
Cao C, Sun J, Zhang D, Guo X, Xie L, Li X, et al. The long intergenic noncoding RNA UFC1, a target of MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to increase levels of beta-catenin in HCC cells. Gastroenterology. 2015;148(2):415-26.e18. https://doi.org/10.1053/j.gastro.2014.10.012(Epub 2014/12/03).
Wu DM, Wang S, Wen X, Han XR, Wang YJ, Shen M, et al. LncRNA SNHG15 acts as a ceRNA to regulate YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma. Cell Death Dis. 2018;9(10):947. https://doi.org/10.1038/s41419-018-0975-1(Epub 2018/09/22).
Ling J, Wang F, Liu C, Dong X, Xue Y, Jia X, et al. FOXO1-regulated lncRNA LINC01197 inhibits pancreatic adenocarcinoma cell proliferation by restraining Wnt/beta-catenin signaling. J Exp Clin Cancer Res. 2019;38(1):179. https://doi.org/10.1186/s13046-019-1174-3(Epub 2019/04/28).
Liu H, Han L, Liu Z, Gao N. Long noncoding RNA MNX1-AS1 contributes to lung cancer progression through the miR-527/BRF2 pathway. J Cell Physiol. 2019;234(8):13843–50. https://doi.org/10.1002/jcp.28064(Epub 2019/01/09).
Sanchez-Mejias A, Tay Y. Competing endogenous RNA networks: tying the essential knots for cancer biology and therapeutics. J Hematol Oncol. 2015;8(1):30. https://doi.org/10.1186/s13045-015-0129-1(Epub 2015/04/19).
Gu J, Wang Y, Wang X, Zhou D, Wang X, Zhou M, et al. Effect of the LncRNA GAS5-MiR-23a-ATG3 axis in regulating autophagy in patients with breast cancer. Cell Physiol Biochem. 2018;48(1):194–207. https://doi.org/10.1159/000491718(Epub 2018/07/17).
Chou ST, Peng HY, Mo KC, Hsu YM, Wu GH, Hsiao JR, et al. MicroRNA-486-3p functions as a tumor suppressor in oral cancer by targeting DDR1. J Exp Clin Cancer Res. 2019;38(1):281. https://doi.org/10.1186/s13046-019-1283-z(Epub 2019/06/30).
Caporali S, Amaro A, Levati L, Alvino E, Lacal PM, Mastroeni S, et al. miR-126-3p down-regulation contributes to dabrafenib acquired resistance in melanoma by up-regulating ADAM9 and VEGF-A. J Exp Clin Cancer Res. 2019;38(1):272. https://doi.org/10.1186/s13046-019-1238-4(Epub 2019/06/23).
Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582(14):1977–86. https://doi.org/10.1016/j.febslet.2008.03.004(Epub 2008/03/18).
Moisan V, Robert NM, Tremblay JJ. Expression of ladybird-like homeobox 2 (LBX2) during ovarian development and folliculogenesis in the mouse. J Mol Histol. 2010;41(4–5):289–94. https://doi.org/10.1007/s10735-010-9291-5(Epub 2010/09/08).
Xie M, Ma T, Xue J, Ma H, Sun M, Zhang Z, et al. The long intergenic non-protein coding RNA 707 promotes proliferation and metastasis of gastric cancer by interacting with mRNA stabilizing protein HuR. Cancer Lett. 2018. https://doi.org/10.1016/j.canlet.2018.11.032(Epub 2018/12/04).
Jia J, Zhan D, Li J, Li Z, Li H, Qian J. The contrary functions of lncRNA HOTAIR/miR-17-5p/PTEN axis and Shenqifuzheng injection on chemosensitivity of gastric cancer cells. J Cell Mol Med. 2018. https://doi.org/10.1111/jcmm.13970(Epub 2018/10/20).
Chen X, Chen Z, Yu S, Nie F, Yan S, Ma P, et al. Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancer. Clin Cancer Res. 2018. https://doi.org/10.1158/1078-0432.ccr-17-2376(Epub 2018/02/02).
Tang LX, Su SF, Wan Q, He P, Xhang Y, Cheng XM. Novel long non-coding RNA LBX2-AS1 indicates poor prognosis and promotes cell proliferation and metastasis through Notch signaling in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2019;23(17):7419–29. https://doi.org/10.26355/eurrev_201909_18851(Epub 2019/09/21).
Cong Z, Diao Y, Xu Y, Li X, Jiang Z, Shao C, et al. Long non-coding RNA linc00665 promotes lung adenocarcinoma progression and functions as ceRNA to regulate AKR1B10-ERK signaling by sponging miR-98. Cell Death Dis. 2019;10(2):84. https://doi.org/10.1038/s41419-019-1361-3(Epub 2019/01/30).
Yang X, Cai JB, Peng R, Wei CY, Lu JC, Gao C, et al. The long noncoding RNA NORAD enhances the TGF-beta pathway to promote hepatocellular carcinoma progression by targeting miR-202-5p. J Cell Physiol. 2019;234(7):12051–60. https://doi.org/10.1002/jcp.27869(Epub 2018/12/12).
Zhang J, Liang Y, Huang X, Guo X, Liu Y, Zhong J, et al. STAT3-induced upregulation of lncRNA MEG3 regulates the growth of cardiac hypertrophy through miR-361-5p/HDAC9 axis. Sci Rep. 2019;9(1):460. https://doi.org/10.1038/s41598-018-36369-1(Epub 2019/01/26).
Acknowledgements
All supports from participants in this research were undeniable.
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval and informed consent
The experiment was permitted by the Research Medical Ethics Committee of the First Affiliated Hospital of Zhengzhou University and the written informed consent was signed by each GC patient. Animal experiment was conducted strictly in light of the protocol approved by the Administrative Panel on Laboratory Animal Care of the First Affiliated Hospital of Zhengzhou University. Ethical approval has been provided by the institute.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhen Yang and Xinhua Dong are co-first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, Z., Dong, X., Pu, M. et al. LBX2-AS1/miR-219a-2-3p/FUS/LBX2 positive feedback loop contributes to the proliferation of gastric cancer. Gastric Cancer 23, 449–463 (2020). https://doi.org/10.1007/s10120-019-01019-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-019-01019-6