Abstract
Carbapenem antibiotics are one of the last-resort agents against multidrug-resistant (MDR) bacteria. The occurrence of carbapenemase-producing Enterobacteriaceae (CPE) in wastewater and aquatic environments is an indication of MDR bacteria in the community. This study evaluated CPE in aquatic environments and compared them to the local hospital isolates in Sweden. Phenotypic and genotypic analyses of antibiotic resistance of environmental and clinical CPE were performed. The relatedness of the isolates and possible clonal dissemination was evaluated using phylogenetic and phyloproteomic analysis. Klebsiella oxytoca carrying carbapenemase genes (blaVIM-1, blaIMP-29) were isolated from wastewater and the recipient river, while K. oxytoca (blaVIM-1) and Klebsiella pneumoniae (blaVIM-1, blaOXA-48, blaNDM-1, blaKPC-3) were isolated from patients at the local clinics or hospital. The K. oxytoca classified as sequence type 172 (ST172) isolated from the river was genotypically related to two clinical isolates recovered from patients. The similarity between environmental and clinical isolates suggests the dispersion of blaVIM-1 producing K. oxytoca ST172 from hospital to aquatic environment and the likelihood of its presence in the community. This is the first report of CPE in aquatic environments in Sweden; therefore, surveillance of aquatic and hospital environments for CPE in other urban areas is important to determine the major transfer routes in order to formulate strategies to prevent the spread of MDR bacteria.
Similar content being viewed by others
Introduction
Infections caused by antibiotic-resistant Enterobacteriaceae are difficult to treat because of their resistance to a broad-spectrum of antibiotics, such as 3rd and 4th generation cephalosporins [1]. Resistance to cephalosporins is primarily due to the presence of extended spectrum β-lactamase (ESBL) enzymes. Carbapenems, including imipenem, meropenem, and ertapenem, are the drugs of choice for treating infections by ESBL producing Enterobacteriaceae; however, resistance to carbapenems has greatly increased worldwide during the last decade [2]. Carbapenemase-producing Enterobacteriaceae (CPE) are an increasing threat to human health due to the limited treatment options for CPE infections such as febrile urinary tract infections (UTI), ventilator-associated pneumonia, abdominal sepsis, and bacteremia [3]. Resistance to carbapenems is primarily attributed to acquired metallo-β-lactamase and serine carbapenemase enzymes (ESBLcarba); however, they may also be combined with other resistance mechanisms such as the loss of outer membrane porins that influence antibiotic uptake [4]. An increase in the number of CPE cases has been reported in hospitals in Sweden and has therefore become notifiable according to the Swedish Communicable Diseases Act since 2012. CPE infections (both infections and carriers of CPE) have increased twofold in Sweden, from 47 cases (0.48/100,000 inhabitants) in 2014 to 115 cases (1.16/100,000 inhabitants) in 2015 [5]. These numbers have further increased to 126 (1.26/100,000 inhabitants) in 2016. The most commonly identified CPE during 2014 was Klebsiella pneumoniae (61%), followed by Escherichia coli (33%) [6].
Klebsiella spp. are ubiquitous in the environment, found in soil, surface water, and on plants [7]. The most important clinical species, K. pneumoniae and Klebsiella oxytoca, are frequently the causative agents of nosocomial infections and have been associated with community-acquired infections such as bacteremia, pneumonia, meningitis, and UTI [8, 9]. The carbapenemase genes, blaOXA-48 (oxacillinase type-48), blaKPC (K. pneumoniae carbapenemase), blaVIM (Verona integron encoded metallo-β-lactamase), blaIMP (imipenemase), and blaNDM (New Delhi metallo-β-lactamase), have been identified in Klebsiella species [10]. Moreover, the NDM-1 gene was first identified in a K. pneumoniae isolated in Örebro, Sweden, from a patient previously hospitalized in New Delhi, India [11]. Klebsiella spp. harboring carbapenemase genes often carry a variety of other antibiotic resistance genes conferring resistance to aminoglycosides and quinolones [12]. Multidrug-resistant (MDR) Klebsiella spp. that include resistance to carbapenems significantly limit the therapeutic options for CPE infections. Colistin and tigecycline are among the remaining drugs effective to treat infections caused by CPE. However, colistin resistance mediated by plasmid-borne mcr-1 and mcr-2 genes as well as tigecycline resistance due to efflux pump overexpression have been recently reported in Enterobacteriaceae [13,14,15,16].
The majority of patients with CPE reported (both carriers or with infections) in Sweden have contracted the CPE abroad [5]. In 71% of CPE cases in 2016, patients were reported to be colonized during international travels and only 27% have acquired CPE from domestic sources [5]. With the increasing occurrence of infections with CPE, there is growing concern over the dissemination of CPE in the natural environment since that may have a direct or indirect effect on human health. Wastewater effluents from hospitals often release clinically relevant bacteria [17], which increase the risk of dissemination of CPE into the environment. Moreover, carbapenemase-harboring bacteria have been reported from non-human sources including animal sewage and river water in both developing and developed countries [17]. Currently, the situation of CPE in aquatic environments and wastewater effluent in Sweden is unknown; therefore, appropriate monitoring of wastewater effluents and environmental waters is ongoing.
The aim of this study was to evaluate the presence of CPE in wastewater and the recipient-river and lake water in Örebro, located in central Sweden, and to investigate the relationship of these bacteria to the clinical isolates obtained from patients in the same city by comparing the genotypic, phenotypic, and phyloproteomic profiles of the isolates.
Methods
Sampling locations
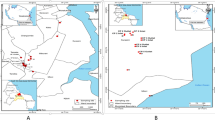
Environmental water samples were collected during October 2015 from Svartån River and Hjälmaren Lake near Örebro, Sweden, a city that reflects urban areas in Nordic countries. The Svartån River flows through Örebro city and is the recipient of the effluent water from Örebro wastewater treatment plant (WWTP) before it flows into Hjälmaren Lake. The WWTP serves a population of 140,000 people and processes an average of 45,000 m3 of wastewater per day (www.orebro.se). The untreated wastewater from Örebro hospital, veterinary clinics, household, and industries is transported via the sewage system to the WWTP. There are no pharmaceutical industries in Örebro city, and most of the agricultural activities are outside of the urban area. Surface water samples from the river were collected from upstream (59° 16′ 03.5″ N, 15° 08′ 49.9″ E) and downstream (59° 16′ 42.5″ N, 15° 15′ 41.5″ E) of Örebro city and WWTP. Surface water samples from Hjälmaren Lake (59° 16′ 40.2″ N, 15° 17′ 31.1″ E) were collected approximately 1 nautical mile downstream of WWTP effluent point. The influent and effluent water from Örebro WWTP was collected to analyze for the presence of CPE. Three separate water samples were collected in sterile 1-litre glass bottles from each location at different times of the day, transported and stored at 4 °C, and analyzed within 24 h.
CPE from a pre-existing collection at Örebro University Hospital were included in the study. The isolates were collected from patients during the years 2008 to 2015.
Bacterial isolation and identification
Water samples were filtered through 0.45 μm polyethylene sulfonate membrane filters (Sartorius Stedim Biotech, Sweden) and placed on selective chromogenic agar medium for carbapenem-resistant bacteria, chromID™ CARBA (bioMérieux, Marcy-l’Etoile, France) and chromID™ OXA-48 (bioMérieux, Marcy-l’Etoile, France). The agar plates were incubated for 18 h at 37 °C, and suspected CPE colonies were picked and streaked onto Chromocult® Coliform Agar (Merck, Darmstadt, Germany) to further identify the isolates as Enterobacteriaceae and to obtain pure cultures.
Isolates were identified using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) with Microflex LT system (Bruker Daltonik GmbH, Bremen, Germany) according to manufacturer’s instructions.
Analysis of carbapenemase production
Putative carbapenemase producers were screened for their susceptibility to meropenem and imipenem using the disk diffusion method according to the EUCAST specifications. The isolates with inhibition zone diameters of ˂ 25 mm and ˂ 23 mm for meropenem (10 μg) and imipenem (10 μg), respectively, were considered presumed carbapenemase producers [18].
Presumed carbapenemase producers were further tested for their carbapenemase production using RAPIDEC® CARBA NP test (BioMérieux, Marcy-l’Etoile, France) according to manufacturer’s instructions [19].
Antibiotic susceptibility testing
The antibiotic susceptibility of isolates was tested for six different classes of antibiotics: beta-lactams (imipenem, meropenem, cefotaxime, ceftazidime, piperacillin-tazobactam), aminoglycosides (gentamicin, amikacin, tobramycin), fluoroquinolone (ciprofloxacin), trimethoprim-sulfamethoxazole, tetracycline (tigecycline), and polymyxin B (colistin). The disk diffusion method was used according to the specifications of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [20]. Minimum inhibitory concentrations were determined using ETEST® (bioMérieux, Marcy-l’Etoile, France) following manufacturer instructions. The plates were incubated at 37 °C for 18 h. EUCAST clinical breakpoint values for zone diameters (mm), and the MIC values were used to categorize the isolates as susceptible (S), intermediate (I), and resistant (R) [21].
DNA isolation and whole genome sequencing
Genomic DNA of Klebsiella spp. was isolated from an overnight culture in nutrient broth (Merck, Germany) using guanidinium thiocyanate-phenol-chloroform extraction method [22]. DNA samples were sent to GATC Biotech (Konstanz, Germany) for whole genome sequencing (WGS). Short paired-end reads of 150 bp were generated using Illumina HiSeq. Quality trimming of the paired-end reads was performed with Trimmomatic version 0.32.3 [23]. De novo genome assembly was performed using SPAdes Genome Assembler version 3.10.1 [24]. The sequences have been submitted to GenBank ID: 451179 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA451179).
Multilocus sequence typing and phylogenetic analysis
MLST was performed using MLST-1.8 Server provided by Center for Genomic Epidemiology [25], and sequence types (STs) were confirmed using SRST2 [26]. For genomic comparisons, we performed core genome SNP analysis and core genome gene-by-gene analysis. To increase the shared sequences for analysis, only K. oxytoca were included in the analysis. Phylogenetic maximum likelihood tree based on core genome SNPs analysis was generated in Parsnp [27] using the default parameters. K. oxytoca JKo3 was selected as the reference genome. Neighbor-joining tree based on core genome MLST (allelic comparison) was performed in SeqSphere+ (Ridom Muenster, Germany) using default parameters, and K. oxytoca JKo3 was selected as the reference genome. Phylogenetic trees were reconstructed in FigTree version 1.4.3 (Institute of Evolutionary Biology, University of Edinburgh). To provide a wider phylogenetic context, we included nine epidemiologically unrelated K. oxytoca genomes in the analysis. The assembled genomes were randomly selected from Ensembl bacterial genome database (bacteria.ensembl.org). The strain names and archive numbers are in Supplementary Table S1.
Antibiotic resistance gene profiling
The antibiotic resistance gene profiling of isolates was performed with oligonucleotide microarray-based assay developed by Alere Technologies [28, 29]. This was complemented with allelic identification of the antibiotic resistance genes (ARGs) from the assembled genomes using Resfinder, server 2.1 [30]. For microarray-based identification of ARGs, fresh cultures from Mueller Hinton agar (BD, LePont de Claix, France) were inoculated to AMIES agar gel tubes (Copan, Brescia, Italy) and sent to Alere Technologies (Germany) for genotyping. The antibiotic resistance gene profiles of both clinical and environmental isolates were analyzed for clinically important carbapenemase genes along with narrow spectrum β-lactamase, ESBL, and other antibiotic resistance genes. The assay included the following carbapenemase genes, blaKPC, blaVIM, blaNDM, blaBIC, blaDIM, blaGES, blaGOB, blaPAM, blaSFH, blaSMB, blaSME, blaSPM, blaTMB, blaGIM, blaIND, blaKHM, as well as blaOXA-23, blaOXA-40, blaOXA-48, blaOXA-50, blaOXA-51, blaOXA-54, and blaOXA-58, and the following ESBL genes, blaCTX-M-1/M-15, blaSHV and blaTEM. In addition, genes conferring resistance to aminoglycosides (e.g., aac, aad, ant2, aphA, strA, strB), quinolones (e.g., qepA, qnrA, qnrB, qnrC, qnrD, qnrS), macrolides (mph, mrx), sulfonamides (sul1, sul2, sul3), and trimethoprim (e.g., dfrA genes) were evaluated (Alere Technologies, http://www.alere-technologies.com). Four genes related to two bacterial toxin-antitoxin systems higB-higA and splT-splA were also included in the assay.
Mass spectra analysis
A loop (approx. 1 μl) of freshly grown bacteria was suspended in ethanol (900 μl, 99.5%), centrifuged at 11,000×g for 2 min, and the pellet was air-dried at room temperature for 3 min. The pellet was incubated in 70% formic acid (20 μl) for 3 min at room temperature to lyse the cells and the released proteins were isolated with acetonitrile (20 μl). The mixture was vortexed and centrifuged at 11,000×g for 2 min. The supernatant was spotted onto a MALDI-TOF MS target plate with Bruker Matrix solution (1 μl). MALDI-TOF MS plates were air-dried at room temperature, and mass spectrometry was performed. Two replicate analyses were performed for each bacterial isolate.
The mass spectra of the bacterial proteins of 2000–20,000 mass-to-charge ratio (m/z) were analyzed using BioNumerics version 7.5 created by Applied Maths NV (http://www.applied-maths.com). Default parameters for strict pre-processing of spectrum data using baseline subtraction, noise elimination, and curve smoothing were selected. Similarity comparison was performed with peak-based Pearson coefficient using default parameters, and phyloproteomic dendrogram was created.
Data availability
The datasets generated during the current study have been deposited in GenBank ID: 451179 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA451179).
Results
Characterization of carbapenemase-producing Klebsiella isolates
A total of 31 putative carbapenem-resistant Gram-negative bacilli were recovered from the WWTP influent (n = 23, 1.87 × 103 CFU/100 ml) and effluent (n = 8, 40 CFU/100 ml) water. From the Svartån River downstream of the WWTP and Hjälmaren Lake, 22 (12 CFU/100 ml) and 16 (8 CFU/100 ml) putative carbapenem-resistant Gram-negative bacilli were recovered, respectively. A total of two isolates were identified as K. oxytoca, one each from Svartån River downstream of the WWTP (4.5% of total carbapenem-resistant Gram-negative bacilli) and influent sewage water (2.5% of total carbapenem-resistant Gram-negative bacilli). Klebsiella spp. were not found in the effluent water from WWTP, Svartån river upstream of the city and Hjälmaren Lake.
In Örebro County, 1132 clinical isolates of ESBL-producing Enterobacteriaceae were reported between 2008 and 2015 [5]. Among those, six (0.53%) isolates were identified as carbapenem-resistant Enterobacteriaceae and were preserved at the Department of Laboratory Medicine, Clinical Microbiology, Örebro University Hospital (Table 1). All the isolates were Klebsiella spp., either K. pneumoniae (n = 4) and K. oxytoca (n = 2), and all were positive for carbapenemase production. It is likely that majority of the patients contracted the CPE abroad since domestic CPE is rare in Sweden.
Genetic analysis of Klebsiella isolates
The antibiotic resistance genes conferring resistance to the different classes of antibiotics were initially determined by an oligonucleotide hybridization assay, and subsequently, the whole genome analysis was used for allelic confirmation of ARGs. The results of these analyses are consolidated in Table 1. A remarkable similarity was observed between one of the K. oxytoca isolates from the river, E1, and two from the hospital (H4 and H6), where H4 showed identical antibiotic resistance gene profiles, while H6 only lacked the strA gene (Table 1). All isolates from the environment and hospital harbored one of five carbapenemase genes. The blaVIM-1 was the most commonly detected carbapenemase gene with 50% isolates carrying this locus. Other carbapenemase genes, blaNDM-1, blaIMP-29, blaKPC-3, and blaOXA-48, were uniquely detected in environmental and clinical isolates (Table 1). Six isolates had one of the metallo-β-lactamase (blaVIM-1, blaNDM-1, blaIMP-29) genes, including the two environmental K. oxytoca isolates from the river (E1) and sewage water (E2). Two clinical K. pneumoniae isolates, H2 and H5, harbored serine-type carbapenemase genes, blaKPC-3 and blaOXA-48, respectively. The genes for two bacterial toxin-antitoxin systems, higB-higA and splT-splA, were not detected in any isolate in this study.
Interestingly, ESBL genes were only detected in K. pneumoniae isolates, while no ESBL genes were detected in any of the K. oxytoca strains (Table 1). K. pneumoniae H1 carried blaCTX-M-15, blaSHV-11 and blaTEM-1, while K. pneumoniae H2 carried two ESBL genes, blaSHV and blaTEM-1. The K. pneumoniae H3 harbored blaCTX-M-9 gene together with blaSHV-28 while H5 harbored blaCTX-M-15 and blaTEM-1 while isolate H3 only harbored blaSHV. All isolates carried antibiotic resistance genes for aminoglycosides and sulfonamides. The common aminoglycoside resistance genes were aac(6′)-Ib, aadA1, strA, and strB. For sulfonamide resistance, sul and dfrA genes were detected. With the exception of the K. oxytoca E2 isolate from the river, quinolone resistance genes were detected in all isolates. Macrolide resistance genes were detected only in K. pneumoniae H1 (mph) and H2 (mph, mrx).
Since K. oxytoca from the river (E1) and hospital (H4) showed identical antibiotic resistance gene profiles according to oligonucleotide hybridization assay, we performed a multilocus sequence typing (MLST) and whole genome phylogenetic analysis to further characterize the isolates. Both of the hospital (H4 and H6) and the environmental (E1) isolates of K. oxytoca belonged to the same sequence type, ST172 (Table 1). Isolate E2 showed a novel allelic profile, which was identified as new sequence type, ST192. All K. pneumoniae isolates showed unique sequence types (Table 1). For phylogenetic analysis, only K. oxytoca were selected while K. pneumoniae were excluded to increase the resolution of core genome analysis. A total of 13 K. oxytoca genomes including isolates from this study (n = 4) and epidemiologically unrelated isolates (n = 9) were subjected to single nucleotide polymorphisms (SNPs) analysis in Parsnp, and phylogenetic tree was constructed based on SNPs comparison (see methods). The Parsnp phylogenetic tree revealed three distinct clusters (Fig. 1a). The K. oxytoca isolates E1, H4, and H6 are together in cluster 3 and are highly related as they share a common genetic ancestor base. The K. oxytoca isolate E2 is placed separately in cluster 2, showing its genetic distance from other K. oxytoca in this study. We further compared homologous coding sequences of isolates using core genome MLST (cgMLST) in SeqSphere+ software. K. oxytoca JKo3 was used as a reference genome, and a set of 3089 common genes in 13 K. oxytoca isolates was included in the analysis. The cgMLST evaluation produced highly similar results to the SNPs analysis (Fig. 1b). Similar to the SNP-based analysis, cgMLST shows that the clinical isolates H4 and H6 and environmental isolate E1 are highly similar and cluster together (cluster 3), while E2 is in a separate cluster (cluster 2).
Phylogenetic relationship of the K. oxytoca isolates from this study compared to other strains. a Phylogenetic maximum-likelihood tree constructed in FigTree on the basis of core genome SNP analysis of using Parsnp. The isolates from the present study are highlighted in red. Scale bar represents genetic distances based on nucleotide substitutions per SNP site. b Allele-based neighbor-joining tree based on 3089 cgMLST target genes showing the clustering of environmental and clinical K. oxytoca strains. cgMLST was performed using K. oxytoca JKo3 as a reference in SeqSphere+. Scale bar represents allelic distance (color figure online)
Multidrug resistance in all the Klebsiella isolates
The eight Klebsiella spp. were tested for susceptibility to six classes of antibiotics (Table 2). K. oxytoca E1 isolate from the river showed intermediate resistance to imipenem while K. oxytoca E2 from incoming wastewater was resistant to imipenem and meropenem but susceptible to ertapenem. Both environmental isolates (E1 and E2) were susceptible to gentamicin, amikacin, colistin, and tigecycline. Among the clinical isolates, both K. oxytoca isolates H4 and H6 were clinically not resistant to all tested carbapenems, aminoglycosides, tetracycline, and polymyxin but showed resistance to cephalosporins, piperacillin-tazobactam, and ciprofloxacin. The K. pneumoniae H1 isolated from a UTI patient was highly resistant to all tested antibiotics except colistin and tigecycline (Table 2), while K. pneumoniae H2 was only susceptible to colistin. All Klebsiella isolates (n = 8) were resistant to ≥ 3 classes of antibiotics and were classified as MDR.
Phyloproteomic comparison of mass spectra of isolates
Strain level comparison between isolates was performed using MALDI-TOF MS and Pearson similarity coefficient analysis. The similarity of mass spectra between each replicate pair was greater than 99% (supplementary Fig. S1), which was used as a threshold value to define related types. K. oxytoca and K. pneumoniae clustered separately with only 12.1% spectral similarity between them (Fig. 2a). The highest similarity of 96% was observed between two clinical K. oxytoca isolates, H4 and H6. A detailed analysis of the spectra was done for the K. oxytoca isolates E1, H4, and H6 to determine their relatedness since they had identical antibiotic resistance genes and H6 only differed by one gene. However, the spectra showed that peaks at mass to charge ratio (m/z) of 2890, 3389, 3418 (Fig. 2b), 5802, and 6625 (Fig. 2c) were absent for E1 but were present in either H4 or H6. The peak corresponding to m/z of 3422 was unique to E1 and absent in H4 and H6. Both environmental isolates of K. oxytoca (E1 and E2) appeared different from the clinical strains with less than 90% similarity (Fig. 2a). Furthermore, a comparison of spectral profiles of environmental K. oxytoca isolates, E1 and E2, revealed 78% similarity and showed a large difference in terms of presence or absence and displacement of peaks (data not shown).
MALDI-TOF mass spectra of the Klebsiella isolates. a Phyloproteomic dendrogram of MALDI-TOF mass spectra of K. pneumoniae and K. oxytoca isolated from Svartån River, incoming sewage to the WWTP and patients from Örebro University Hospital. b, c Zoom into spectral regions with differences between K. oxytoca isolates from the river (E1) and hospital (H4 and H6), based on the presence and absence of ions of specific mass to charge ratio. Five major differences between the strains are shown in ellipses
Discussion
In the present study, carbapenemase-producing K. oxytoca were isolated from sewage and river waters running through Örebro city in central Sweden. To the best of our knowledge, this was the first CPE isolated from the environment in Sweden, thus prior to 2015, no isolates were reported. This is in contrast to other European countries that have reported health-care associated CPE since 2011 [31]. Only two carbapenemase-producing Klebsiella isolates were recovered from the environment, indicating that the levels of CPE in environmental waters remain very low. However, the presence of these resistant bacteria in the river, even at low levels, remains a concern. Although carbapenemase-producing K. oxytoca were found in Svartån River, they were not detected in the downstream lake water, which suggests that CPE may be present at even lower levels or completely absent in the lake due to the increased dilution or die-off under environmental conditions. Environmental K. oxytoca isolates, E1 and E2, harbored blaVIM-1 and blaIMP-29, respectively, and also showed phenotypic resistance to either one or two carbapenem antibiotics (Table 2). These carbapenemase genes are the most commonly identified worldwide and blaVIM-1 is predominant in Europe, while blaIMP alleles are prevalent in Asia [32]. As none of the isolates from patients in Örebro University Hospital were identified with blaIMP-29, this implies that the source of CPE in the wastewater most likely originate from the community.
Only six cases of CPE infections were reported in Örebro County between 2008 and 2015, and the majority of these patients had likely contracted their resistant bacteria from abroad rather than domestic acquisition in the Swedish hospital environment. The clinical K. pneumoniae isolates H1 and H2 harbored blaNDM-1 and blaKPC-3, respectively, and the blaNDM type metallo-β-lactamases are highly prevalent in the Indian sub-continent [33, 34], while blaKPC carbapenemases have worldwide prevalence [35,36,37,38]. These isolates harbored antibiotic resistance genes for a broad range of antibiotics as well as showed functional resistance to most of the antibiotics tested in this study. The K. pneumoniae H1 co-harbored other ESBL genes such as blaCTX-M-15, which is frequently detected in blaNDM-1-positive Enterobacteriaceae [39] and also the predominant ESBL type in Örebro County [40]. Strains H4 and H6 only carried blaVIM-1, and these had high genome similarity to the strain from river (E1) indicating that this isolate might be widespread in the community.
Resistance to carbapenems was due to the presence of functional carbapenemase genes; however, other mechanisms that were not analyzed in this study could also contribute the final phenotype. These mechanisms include the overexpression of efflux-pumps and downregulation or modification of outer-membrane proteins such as ompK and phoE [41]. Most carbapenemase genes are found on mobile plasmids that can be exchanged between the members of Enterobacteriaceae through horizontal gene transfer [42]. Some plasmids carry carbapenemase genes along with toxin-antitoxin systems that increase plasmid prevalence by post-segregational killing of daughter cells that lack the plasmid [43]. Thus, antibiotic selection pressure is not always necessary to maintain antibiotic resistance plasmids in a bacterial population. The recipient WWTP of the hospital and municipal wastewater is a major source of antibiotic-resistant Enterobacteriaceae, and the treatment process often reduces but does not eliminate these organisms [44]. Complex and nutrient-rich microbial environment in the WWTP may provide ideal conditions for the expansion of antibiotic resistance into natural environments [45] and may help to disseminate plasmids with toxin-antitoxin systems in a bacterial population without any selection pressure. It is interesting to note that our strains did not have the higB-higA and splT-splA toxin-antitoxin genes. However, there is a possibility of the presence of other toxin-antitoxin systems that were not analyzed in this study.
In countries with low to middle-income economies, untreated wastewaters from households and pharmaceutical industries are discharged into natural environments without prior treatment, and this has been shown to increase antibiotic-resistant bacteria in the environment [46]. In Sweden, however, the regulations for wastewater disposal have been strict since the approval of the Environmental Protection Act in 1969. Our study revealed the presence of CPE in the Swedish aquatic environment, albeit at low levels, regardless of the stringent regulations for antibiotic use. Although, the study is based on the comparison of a small set of strains, the presence of related CPE in both hospital and associated river and lake environments is an important observation. This shows the possibility of transmission of CPE from hospital to aquatic environment since the effluent wastewater from hospital is transported through the drainage system into the WWTP without prior treatment. In addition to the hospital, the community and veterinary clinics are other potential sources of CPE that may reach the aquatic environment, with the WWTP being the distributor. The clinical isolates carrying carbapenemase genes were most probably acquired from abroad and not generated at the hospital, suggesting that alternate strategies must be employed to avoid the further environmental contamination and transmission of MDR in the community. Although WWTP plays an important role to minimize the organic pollution from the wastewater, they can be ultimate points of dissemination for MDR bacteria into natural environments [47]. Therefore, WWTP must apply processes for eradicating bacteria, including MDR bacteria from wastewater before discharging into the environment. Thus, attempts to reduce the spread of CPE to the aquatic environment in Sweden and surrounding countries are of vital importance since the presence of CPE in these niches is still rare.
References
Cao X, Cavaco LM, Lv Y, Li Y, Zheng B, Wang P, Hasman H, Liu Y, Aarestrup FM (2011) Molecular characterization and antimicrobial susceptibility testing of Escherichia coli isolates from patients with urinary tract infections in 20 Chinese hospitals. J Clin Microbiol 49(7):2496–2501. https://doi.org/10.1128/JCM.02503-10
Braykov NP, Eber MR, Klein EY, Morgan DJ, Laxminarayan R (2013) Trends in resistance to carbapenems and third-generation cephalosporins among clinical isolates of Klebsiella pneumoniae in the United States, 1999–2010. Infect Control Hosp Epidemiol 34(3):259–268. https://doi.org/10.1086/669523
Tzouvelekis LS, Markogiannakis A, Piperaki E, Souli M, Daikos GL (2014) Treating infections caused by carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect 20(9):862–872. https://doi.org/10.1111/1469-0691.12697
Yang D, Guo Y, Zhang Z (2009) Combined porin loss and extended spectrum β-lactamase production is associated with an increasing imipenem minimal inhibitory concentration in clinical Klebsiella pneumoniae strains. Curr Microbiol 58(4):366–370. https://doi.org/10.1007/s00284-009-9364-4
Folkhälsomyndigheten (2016) Yearly statistics of ESBLcarba producing Enterobacteriaceae in Sweden by Public Health Agency
SWEDRES-SVARM (2014) Consumption of antibiotics and occurrence of antibiotic resistance in Sweden. Solna/Uppsala ISSN 1650–6332
Bagley ST (1985) Habitat association of Klebsiella species. Infect Control 6(2):52–58
Lin YT, Jeng YY, Chen TL, Fung CP (2010) Bacteremic community-acquired pneumonia due to Klebsiella pneumoniae: clinical and microbiological characteristics in Taiwan, 2001–2008. BMC Infect Dis 10:307. https://doi.org/10.1186/1471-2334-10-307
Podschun R, Ullmann U (1998) Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 11(4):589–603
Patel G, Bonomo RA (2011) Status report on carbapenemases: challenges and prospects. Expert Rev Anti-Infect Ther 9(5):555–570. https://doi.org/10.1586/eri.11.28
Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR (2009) Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53(12):5046–5054. https://doi.org/10.1128/aac.00774-09
Uz Zaman T, Aldrees M, Al Johani SM, Alrodayyan M, Aldughashem FA, Balkhy HH (2014) Multi-drug carbapenem-resistant Klebsiella pneumoniae infection carrying the OXA-48 gene and showing variations in outer membrane protein 36 causing an outbreak in a tertiary care hospital in Riyadh, Saudi Arabia. Int J Infect Dis 28:186–192. https://doi.org/10.1016/j.ijid.2014.05.021
Yao X, Doi Y, Zeng L, Lv L, Liu J-H (2016) Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis 16(3):288–289. https://doi.org/10.1016/S1473-3099(16)00057-8
Marchaim D, Chopra T, Pogue JM, Perez F, Hujer AM, Rudin S, Endimiani A, Navon-Venezia S, Hothi J, Slim J, Blunden C, Shango M, Lephart PR, Salimnia H, Reid D, Moshos J, Hafeez W, Bheemreddy S, Chen T-Y, Dhar S, Bonomo RA, Kaye KS (2011) Outbreak of colistin-resistant, carbapenem-resistant Klebsiella pneumoniae in Metropolitan Detroit, Michigan. Antimicrob Agents Chemother 55(2):593–599. https://doi.org/10.1128/aac.01020-10
Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S (2016) Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21(27). https://doi.org/10.2807/1560-7917.es.2016.21.27.30280
Pournaras S, Koumaki V, Spanakis N, Gennimata V, Tsakris A (2016) Current perspectives on tigecycline resistance in Enterobacteriaceae: susceptibility testing issues and mechanisms of resistance. Int J Antimicrob Agents 48(1):11–18. https://doi.org/10.1016/j.ijantimicag.2016.04.017
Woodford N, Wareham DW, Guerra B, Teale C (2014) Carbapenemase-producing Enterobacteriaceae and non-Enterobacteriaceae from animals and the environment: an emerging public health risk of our own making? J Antimicrob Chemother 69(2):287–291. https://doi.org/10.1093/jac/dkt392
EUCAST (2013) Guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance
Poirel L, Nordmann P (2015) Rapidec Carba NP test for rapid detection of carbapenemase producers. J Clin Microbiol 53(9):3003–3008. https://doi.org/10.1128/jcm.00977-15
EUCAST (2013) Disk diffusion test methodology
EUCAST (2016) Breakpoint tables for interpretation of MICs and zone diameters
Lemarchand K, Berthiaume F, Maynard C, Harel J, Payment P, Bayardelle P, Masson L, Brousseau R (2005) Optimization of microbial DNA extraction and purification from raw wastewater samples for downstream pathogen detection by microarrays. J Microbiol Methods 63(2):115–126. https://doi.org/10.1016/j.mimet.2005.02.021
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477. https://doi.org/10.1089/cmb.2012.0021
Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O (2012) Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50(4):1355–1361. https://doi.org/10.1128/jcm.06094-11
Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE (2014) SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6(11):90. https://doi.org/10.1186/s13073-014-0090-6
Treangen TJ, Ondov BD, Koren S, Phillippy AM (2014) The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15(11):524. https://doi.org/10.1186/s13059-014-0524-x
Braun SD, Monecke S, Thürmer A, Ruppelt A, Makarewicz O, Pletz M, Reißig A, Slickers P, Ehricht R (2014) Rapid identification of carbapenemase genes in gram-negative bacteria with an oligonucleotide microarray-based assay. PLoS One 9(7):e102232. https://doi.org/10.1371/journal.pone.0102232
Braun SD, Dorneanu OS, Vremera T, Reissig A, Monecke S, Ehricht R (2016) Carbapenemase-producing Enterobacteriaceae: a 2-year surveillance in a hospital in Iasi, Romania. Future Microbiol 11:391–401. https://doi.org/10.2217/fmb.15.148
Kleinheinz KA, Joensen KG, Larsen MV (2014) Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage 4:e27943. https://doi.org/10.4161/bact.27943
Albiger B, Glasner C, Struelens MJ, Grundmann H, Monnet DL (2015) Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro Surveill 20(45). https://doi.org/10.2807/1560-7917.es.2015.20.45.30062
Tato M, Coque TM, Baquero F, Cantón R (2010) Dispersal of carbapenemase blaVIM-1 gene associated with different Tn402 variants, mercury transposons, and conjugative plasmids in Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 54(1):320–327. https://doi.org/10.1128/aac.00783-09
Perry JD, Naqvi SH, Mirza IA, Alizai SA, Hussain A, Ghirardi S, Orenga S, Wilkinson K, Woodford N, Zhang J, Livermore DM, Abbasi SA, Raza MW (2011) Prevalence of faecal carriage of Enterobacteriaceae with NDM-1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media. J Antimicrob Chemother 66(10):2288–2294. https://doi.org/10.1093/jac/dkr299
Bharadwaj R, Joshi S, Dohe V, Gaikwad V, Kulkarni G, Shouche Y (2012) Prevalence of New Delhi metallo-beta-lactamase (NDM-1)-positive bacteria in a tertiary care centre in Pune, India. Int J Antimicrob Agents 39(3):265–266. https://doi.org/10.1016/j.ijantimicag.2011.09.027
Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG (2009) Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53(8):3365–3370. https://doi.org/10.1128/aac.00126-09
Sheppard AE, Stoesser N, Wilson DJ, Sebra R, Kasarskis A, Anson LW, Giess A, Pankhurst LJ, Vaughan A, Grim CJ, Cox HL, Yeh AJ, Sifri CD, Walker AS, Peto TE, Crook DW, Mathers AJ (2016) Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob Agents Chemother 60(6):3767–3778. https://doi.org/10.1128/aac.00464-16
Stoesser N, Sheppard AE, Peirano G, Anson LW, Pankhurst L, Sebra R, Phan HTT, Kasarskis A, Mathers AJ, Peto TEA, Bradford P, Motyl MR, Walker AS, Crook DW, Pitout JD (2017) Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli. Sci Rep 7(1):5917. https://doi.org/10.1038/s41598-017-06256-2
Nordmann P, Cuzon G, Naas T (2009) The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 9(4):228–236. https://doi.org/10.1016/S1473-3099(09)70054-4
Poirel L, Dortet L, Bernabeu S, Nordmann P (2011) Genetic features of bla(NDM-1)-positive Enterobacteriaceae. Antimicrob Agents Chemother 55(11):5403–5407. https://doi.org/10.1128/AAC.00585-11
Önnberg A, Söderquist B, Persson K, Mölling P (2014) Characterization of CTX-M-producing Escherichia coli by repetitive sequence-based PCR and real-time PCR-based replicon typing of CTX-M-15 plasmids. APMIS 122(11):1136–1143. https://doi.org/10.1111/apm.12270
Yigit H, Marie Queenan A, Kamile Rasheed J, Biddle JW, Doménech-Sánchez A, Alberti S, Bush K, Tenover FC (2004) Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing -lactamase KPC-2. 47. https://doi.org/10.1128/AAC.47.12.3881-3889.2003
Goren MG, Carmeli Y, Schwaber MJ, Chmelnitsky I, Schechner V, Navon-Venezia S (2010) Transfer of Carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerg Infect Dis 16(6):1014–1017. https://doi.org/10.3201/eid1606.091671
Mosqueda N, Gato E, Roca I, Lopez M, de Alegria CR, Fernandez Cuenca F, Martinez-Martinez L, Pachon J, Cisneros JM, Rodriguez-Bano J, Pascual A, Vila J, Bou G, Tomas M (2014) Characterization of plasmids carrying the blaOXA-24/40 carbapenemase gene and the genes encoding the AbkA/AbkB proteins of a toxin/antitoxin system. J Antimicrob Chemother 69(10):2629–2633. https://doi.org/10.1093/jac/dku179
Aali R, Nikaeen M, Khanahmad H, Hassanzadeh A (2014) Monitoring and comparison of antibiotic resistant bacteria and their resistance genes in municipal and hospital wastewaters. Int J Prev Med 5(7):887–894
Kummerer K (2009) Antibiotics in the aquatic environment—a review—part II. Chemosphere 75(4):435–441. https://doi.org/10.1016/j.chemosphere.2008.12.006
Marathe N, Pal C, Gaikwad S, Jonsson V, Kristiansson E, Larsson J (2017) Untreated urban waste contaminates Indian river sediments with resistance genes to last resort antibiotics. 124. https://doi.org/10.1016/j.watres.2017.07.060
Rutgersson C, Fick J, Marathe N, Kristiansson E, Janzon A, Angelin M, Johansson A, Shouche Y, Flach CF, Larsson DG (2014) Fluoroquinolones and qnr genes in sediment, water, soil, and human fecal flora in an environment polluted by manufacturing discharges. Environ Sci Technol 48(14):7825–7832. https://doi.org/10.1021/es501452a
Acknowledgments
The authors thank Ellinor Rapp and Ines Engelmann for technical assistance.
Funding
Svenska Forskningsrådet Formas (219-2014-837) (JJ) and Nyckelfonden at Örebro University Hospital (BS).
Author information
Authors and Affiliations
Contributions
J.J. and B.S. supervised the project and together with F.A.H. designed the study, analyzed, and interpreted the data. F.A.H. collected the data and wrote the initial draft of the manuscript. B.H. contributed to analysis of the clinical strains and MALDI-TOF MS data, and R.E. provided the gene array analysis of all the strains. J.J., B.S., and R.E. critically revised the manuscript, and all authors contributed to the writing and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
R.E. is employed at Abbott (ALERE Technologies GmbH). This has not influenced the study.
Ethical approval
For this type of study, formal consent is not required.
Electronic supplementary material
ESM 1
(DOCX 3.46 mb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Khan, F.A., Hellmark, B., Ehricht, R. et al. Related carbapenemase-producing Klebsiella isolates detected in both a hospital and associated aquatic environment in Sweden. Eur J Clin Microbiol Infect Dis 37, 2241–2251 (2018). https://doi.org/10.1007/s10096-018-3365-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-018-3365-9