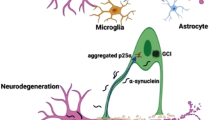
Abstract
Background
Parkinson’s disease (PD) ranks as the second most prevalent neurodegenerative disorder globally, and its incidence is rapidly rising. The diagnosis of PD relies on clinical characteristics. Although current treatments aim to alleviate symptoms, they do not effectively halt the disease’s progression. Early detection and intervention hold immense importance. This study aimed to establish a new PD diagnostic model.
Methods
Data from a public database were adopted for the construction and validation of a PD diagnostic model with random forest and artificial neural network models. The CIBERSORT platform was applied for the evaluation of immune cell infiltration in PD. Quantitative real-time PCR was performed to verify the accuracy and reliability of the bioinformatics analysis results.
Results
Leveraging existing gene expression data from the Gene Expression Omnibus (GEO) database, we sifted through differentially expressed genes (DEGs) in PD and identified 30 crucial genes through a random forest classifier. Furthermore, we successfully designed a novel PD diagnostic model using an artificial neural network and verified its diagnostic efficacy using publicly available datasets. Our research also suggests that mast cells may play a significant role in the onset and progression of PD.
Conclusion
This work developed a new PD diagnostic model with machine learning techniques and suggested the immune cells as a potential target for PD therapy.






Similar content being viewed by others
Data availability
All data relevant to the study are included in the article or are uploaded as Supplementary Information. The data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
(2014) Alzheimer’s disease facts and figures. Alzheimer’s Dementia: J Alzheimer’s Assoc 10:e47-92.https://doi.org/10.1016/j.jalz.2014.02.001
Tysnes OB, Storstein A (2017) Epidemiology of Parkinson’s disease. J Neural Transm (Vienna, Austria: 1996) 124:901–905. https://doi.org/10.1007/s00702-017-1686-y
Yang W et al (2020) Current and projected future economic burden of Parkinson’s disease in the U.S. NPJ Parkinson’s Dis 6:15. https://doi.org/10.1038/s41531-020-0117-1
Jankovic J, Tan EK (2020) Parkinson’s disease: etiopathogenesis and treatment. J Neurol Neurosurg Psychiatry 91:795–808. https://doi.org/10.1136/jnnp-2019-322338
Bendor JT, Logan TP, Edwards RH (2013) The function of α-synuclein. Neuron 79:1044–1066. https://doi.org/10.1016/j.neuron.2013.09.004
Killinger BA, Kordower JH (2019) Spreading of alpha-synuclein - relevant or epiphenomenon? J Neurochem 150:605–611. https://doi.org/10.1111/jnc.14779
Dickson DW et al (2009) Neuropathological assessment of Parkinson’s disease: refining the diagnostic criteria. Lancet Neurol 8:1150–1157. https://doi.org/10.1016/s1474-4422(09)70238-8
Iwai A et al (1995) The precursor protein of non-A beta component of Alzheimer’s disease amyloid is a presynaptic protein of the central nervous system. Neuron 14:467–475. https://doi.org/10.1016/0896-6273(95)90302-x
Ahmadi SA et al (2020) Analyzing the co-localization of substantia nigra hyper-echogenicities and iron accumulation in Parkinson’s disease: a multi-modal atlas study with transcranial ultrasound and MRI. NeuroImage Clin 26:102185. https://doi.org/10.1016/j.nicl.2020.102185
Obeso JA et al (2017) Past, present, and future of Parkinson’s disease: a special essay on the 200th Anniversary of the Shaking Palsy. Mov Disord: Off J Mov Disord Soc 32:1264–1310. https://doi.org/10.1002/mds.27115
Del Tredici K, Braak H (2012) Lewy pathology and neurodegeneration in premotor Parkinson’s disease. Mov Disord: Off J Mov Disord Soc 27:597–607. https://doi.org/10.1002/mds.24921
Lenka A, Padmakumar C, Pal PK (2017) Treatment of older Parkinson’s disease. Int Rev Neurobiol 132:381–405. https://doi.org/10.1016/bs.irn.2017.01.005
Braak H, Ghebremedhin E, Rüb U, Bratzke H, Del Tredici K (2004) Stages in the development of Parkinson’s disease-related pathology. Cell Tissue Res 318:121–134. https://doi.org/10.1007/s00441-004-0956-9
Fox SH et al (2018) International Parkinson and movement disorder society evidence-based medicine review: update on treatments for the motor symptoms of Parkinson’s disease. Mov Disord: Off J Mov Disord Soc 33:1248–1266. https://doi.org/10.1002/mds.27372
Reich SG, Savitt JM (2019) Parkinson’s disease. Med Clin North Am 103:337–350. https://doi.org/10.1016/j.mcna.2018.10.014
Surmeier DJ, Obeso JA, Halliday GM (2017) Selective neuronal vulnerability in Parkinson disease. Nat Rev Neurosci 18:101–113. https://doi.org/10.1038/nrn.2016.178
Titova N, Padmakumar C, Lewis SJG, Chaudhuri KR (2017) Parkinson’s: a syndrome rather than a disease? J Neural Transm (Vienna, Austria: 1996) 124:907–914. https://doi.org/10.1007/s00702-016-1667-6
Greenland JC, Williams-Gray CH, Barker RA (2019) The clinical heterogeneity of Parkinson’s disease and its therapeutic implications. Eur J Neurosci 49:328–338. https://doi.org/10.1111/ejn.14094
Clark LN, Louis ED (2018) Essential tremor. Handbook Clin Neurol 147:229–239. https://doi.org/10.1016/b978-0-444-63233-3.00015-4
Deng H, Wang P, Jankovic J (2018) The genetics of Parkinson disease. Ageing Res Rev 42:72–85. https://doi.org/10.1016/j.arr.2017.12.007
Blauwendraat C, Nalls MA, Singleton AB (2020) The genetic architecture of Parkinson’s disease. Lancet Neurol 19:170–178. https://doi.org/10.1016/s1474-4422(19)30287-x
Saberi-Karimian M et al (2021) Potential value and impact of data mining and machine learning in clinical diagnostics. Crit Rev Clin Lab Sci 58:275–296. https://doi.org/10.1080/10408363.2020.1857681
Tai AMY et al (2019) Machine learning and big data: implications for disease modeling and therapeutic discovery in psychiatry. Artif Intell Med 99:101704. https://doi.org/10.1016/j.artmed.2019.101704
Breiman L (2001) Random forests. Mach Learn 45:5–32. https://doi.org/10.1023/A:1010933404324
Han S, Kim H, Lee Y-S (2020) Double random forest. Mach Learn 109:1569–1586. https://doi.org/10.1007/s10994-020-05889-1
Thiessen ED (2017) What’s statistical about learning? Insights from modelling statistical learning as a set of memory processes. Phil Trans R Soc London. Series B, Biol Sci 372. https://doi.org/10.1098/rstb.2016.0056
Qi Y (2012) Random forest for bioinformatics. Ensemble Mach Learn: Methods Appl. https://doi.org/10.1007/978-1-4419-9326-7_11
Cohen Y et al (2022) Recent advances at the interface of neuroscience and artificial neural networks. J Neurosci: Off J Soc Neurosci 42:8514–8523. https://doi.org/10.1523/jneurosci.1503-22.2022
Khan J et al (2001) Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med 7:673–679. https://doi.org/10.1038/89044
Nayarisseri A et al (2021) Artificial intelligence, big data and machine learning approaches in precision medicine & drug discovery. Curr Drug Targets 22:631–655. https://doi.org/10.2174/1389450122999210104205732
Durrenberger PF et al (2015) Common mechanisms in neurodegeneration and neuroinflammation: a BrainNet Europe gene expression microarray study. J Neural Transm (Vienna, Austria: 1996) 122:1055–1068. https://doi.org/10.1007/s00702-014-1293-0
Zheng B et al (2010) PGC-1α, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med 2:52ra73. https://doi.org/10.1126/scitranslmed.3001059
Durrenberger PF et al (2012) Selection of novel reference genes for use in the human central nervous system: a BrainNet Europe Study. Acta Neuropathol 124:893–903. https://doi.org/10.1007/s00401-012-1027-z
Huang DW et al (2007) DAVID Bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 35:W169-175. https://doi.org/10.1093/nar/gkm415
Huang DW et al (2007) The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8:R183. https://doi.org/10.1186/gb-2007-8-9-r183
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. Omics: J Integr Biol 16:284–287. https://doi.org/10.1089/omi.2011.0118
Liu Y, Zhao H (2017) Variable importance-weighted random forests. Quant Biol (Beijing, China) 5:338–351
Beck MW (2018) NeuralNetTools: visualization and analysis tools for neural networks. J Stat Softw 85:1–20. https://doi.org/10.18637/jss.v085.i11
Robin X et al (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12:77. https://doi.org/10.1186/1471-2105-12-77
Michael Friendly YU (2002) Corrgrams: exploratory displays for correlation matrices. Am Stat 56:316–324
Surmeier DJ (2018) Determinants of dopaminergic neuron loss in Parkinson’s disease. FEBS J 285:3657–3668. https://doi.org/10.1111/febs.14607
Blauwendraat C et al (2019) Parkinson’s disease age at onset genome-wide association study: defining heritability, genetic loci, and α-synuclein mechanisms. Mov Disord: Off J Mov Disord Soc 34:866–875. https://doi.org/10.1002/mds.27659
Chen X, Cao W, Zhuang Y, Chen S, Li X (2021) Integrative analysis of potential biomarkers and immune cell infiltration in Parkinson’s disease. Brain Res Bull 177:53–63. https://doi.org/10.1016/j.brainresbull.2021.09.010
Bae JR, Kim SH (2017) Synapses in neurodegenerative diseases. BMB Rep 50:237–246. https://doi.org/10.5483/bmbrep.2017.50.5.038
Goedert M (2001) Alpha-synuclein and neurodegenerative diseases. Nat Rev Neurosci 2:492–501. https://doi.org/10.1038/35081564
Burré J (2015) The synaptic function of α-synuclein. J Parkinson’s Dis 5:699–713. https://doi.org/10.3233/jpd-150642
Gitler AD et al (2008) The Parkinson’s disease protein alpha-synuclein disrupts cellular Rab homeostasis. Proc Natl Acad Sci USA 105:145-150.https://doi.org/10.1073/pnas.0710685105
Oorschot DE (1996) Total number of neurons in the neostriatal, pallidal, subthalamic, and substantia nigral nuclei of the rat basal ganglia: a stereological study using the cavalieri and optical disector methods. J Comp Neurol 366:580–599. https://doi.org/10.1002/(sici)1096-9861(19960318)366:4%3c580::Aid-cne3%3e3.0.Co;2-0
Roberts RC, Force M, Kung L (2002) Dopaminergic synapses in the matrix of the ventrolateral striatum after chronic haloperidol treatment. Synapse (New York, N.Y.) 45:78–85. https://doi.org/10.1002/syn.10081
Bridi JC, Hirth F (2018) Mechanisms of α-synuclein induced synaptopathy in Parkinson’s disease. Front Neurosci 12:80. https://doi.org/10.3389/fnins.2018.00080
Busch DJ et al (2014) Acute increase of α-synuclein inhibits synaptic vesicle recycling evoked during intense stimulation. Mol Biol Cell 25:3926–3941. https://doi.org/10.1091/mbc.E14-02-0708
Janezic S et al (2013) Deficits in dopaminergic transmission precede neuron loss and dysfunction in a new Parkinson model. Proc Natl Acad Sci USAm 110:E4016-4025.https://doi.org/10.1073/pnas.1309143110
Lee H, James WS, Cowley SA (2017) LRRK2 in peripheral and central nervous system innate immunity: its link to Parkinson’s disease. Biochem Soc Trans 45:131–139. https://doi.org/10.1042/bst20160262
Sweeney MD, Sagare AP, Zlokovic BV (2018) Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat Rev Neurol 14:133–150. https://doi.org/10.1038/nrneurol.2017.188
Yu X, Yao JY, He J, Tian JW (2015) Protection of MPTP-induced neuroinflammation and neurodegeneration by rotigotine-loaded microspheres. Life Sci 124:136–143. https://doi.org/10.1016/j.lfs.2015.01.014
Kempuraj D et al (2019) Mast cell proteases activate astrocytes and glia-neurons and release interleukin-33 by activating p38 and ERK1/2 MAPKs and NF-κB. Mol Neurobiol 56:1681–1693. https://doi.org/10.1007/s12035-018-1177-7
Bergot AS et al (2014) HPV16-E7 expression in squamous epithelium creates a local immune suppressive environment via CCL2- and CCL5- mediated recruitment of mast cells. PLoS Pathog 10:e1004466. https://doi.org/10.1371/journal.ppat.1004466
Hong GU, Kim NG, Jeoung D, Ro JY (2013) Anti-CD40 Ab- or 8-oxo-dG-enhanced Treg cells reduce development of experimental autoimmune encephalomyelitis via down-regulating migration and activation of mast cells. J Neuroimmunol 260:60–73. https://doi.org/10.1016/j.jneuroim.2013.04.002
Hong GU, Cho JW, Kim SY, Shin JH, Ro JY (2018) Inflammatory mediators resulting from transglutaminase 2 expressed in mast cells contribute to the development of Parkinson’s disease in a mouse model. Toxicol Appl Pharmacol 358:10–22. https://doi.org/10.1016/j.taap.2018.09.003
Zhang C, Jiang H, Wang P, Liu H, Sun X (2017) Transcription factor NF-kappa B represses ANT1 transcription and leads to mitochondrial dysfunctions. Sci Rep 7:44708. https://doi.org/10.1038/srep44708
Brochard V et al (2009) Infiltration of CD4+ lymphocytes into the brain contributes to neurodegeneration in a mouse model of Parkinson disease. J Clin Investig 119:182–192. https://doi.org/10.1172/jci36470
Skaper SD, Facci L, Giusti P (2014) Mast cells, glia and neuroinflammation: partners in crime? Immunology 141:314–327. https://doi.org/10.1111/imm.12170
Kempuraj D et al (2018) Cross-talk between glia, neurons and mast cells in neuroinflammation associated with Parkinson’s disease. J Neuroimmune Pharmacol: Off J Soc NeuroImmune Pharmacol 13:100–112. https://doi.org/10.1007/s11481-017-9766-1
Hendriksen E, van Bergeijk D, Oosting RS, Redegeld FA (2017) Mast cells in neuroinflammation and brain disorders. Neurosci Biobehav Rev 79:119–133. https://doi.org/10.1016/j.neubiorev.2017.05.001
Funding
This work is funded by Scientific Research Project of Hunan Provincial Health Commission (D202303077877), and this work was supported by the China Postdoctoral Science Foundation (No.2022M713535), the Provincial Natural Science Foundation of Hunan (No.2023JJ41005).
Author information
Authors and Affiliations
Contributions
Shucai Xie designed the study. Pei Peng and Xingcheng Dong collated the data. Ji Liang and Shucai Xie carried out data analyses and produced the initial draft of the manuscript. Shucai Xie and Ji Liang contributed to drafting the manuscript. Shucai Xie and Junxing Yuan conducted experiments. All authors have read and approved the final submitted manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xie, S., Peng, P., Dong, X. et al. Novel gene signatures predicting and immune infiltration analysis in Parkinson’s disease: based on combining random forest with artificial neural network. Neurol Sci 45, 2681–2696 (2024). https://doi.org/10.1007/s10072-023-07299-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-023-07299-2