Abstract
We report on a boy with myoclonus-dystonia (M-D), language delay, and malformative anomalies. Genetic investigations allowed the identification of an apparently balanced de novo reciprocal translocation, t(7;9)(q21;p23). Breakpoint-region mapping using fluorescent in situ hybridization (FISH) analysis of bacterial artificial chromosome (BAC) clone probes identified microdeletions of 3.7 and 5.2 Mb within 7q21 and 9p23 breakpoint regions, respectively. Genotyping with microsatellite markers showed that deletions originated from paternal alleles. The deleted region on chromosome 7q21 includes a large imprinted gene cluster. SGCE and PEG10 are two maternally imprinted genes. SGCE mutations are implicated in M-D. In our case, M-D is due to deletion of the paternal allele of the SGCE gene. PEG10 is strongly expressed in the placenta and is essential for embryo development. Prenatal growth retardation identified in the patient may be due to deletion of the paternal allele of the PEG10 gene. Other genes in the deleted region on chromosome 7 are not imprinted. Nevertheless, a phenotype can be due to haploinsufficiency of these genes. KRIT1 is implicated in familial forms of cerebral cavernous malformations, and COL1A2 may be implicated in very mild forms of osteogenesis imperfecta. The deleted region on chromosome 9 overlaps with the candidate region for monosomy 9p syndrome. The proband shows dysmorphic features compatible with monosomy 9p syndrome, without mental impairment. These results emphasize that the phenotypic abnormalities of apparently balanced de novo translocations can be due to cryptic deletions and that the precise mapping of these aneusomies may improve clinical management.
Similar content being viewed by others
Introduction
Myoclonus-dystonia (M-D, DYT11, OMIM 159900) is a dystonia plus syndrome. It begins in childhood or adolescence and is characterized by brief myoclonic muscle jerks frequently accompanied by mild dystonia affecting the arms, trunk, or bulbar muscles (Nardocci et al. 2008). Optional diagnostic criteria are a positive response of symptoms to alcohol, and various personality disorders and psychiatric disturbances. Heterozygous loss-of-function mutations in the gene encoding SGCE, located on chromosome 7q21, have been identified in several cases (Zimprich et al. 2001). Most patients inherited the mutant allele from their father, suggesting a maternal imprinting (Muller et al. 2002; Grabowski et al. 2003). In general, large genomic deletions are not as common as single-base alterations; however, they contribute significantly to the proportion of human disease. Structural anomalies involving human chromosomes associated with abnormal phenotype are very informative because they may contribute to identify genes involved in the phenotype. In this study, we report on a boy with M-D and malformative anomalies associated with apparently balanced de novo reciprocal translocation between chromosomes 7 and 9. Breakpoint-region mapping allowed us to identify cryptic deletions, which could participate in the pathological phenotype through abnormal expression of dosage-sensitive genes.
Clinical report
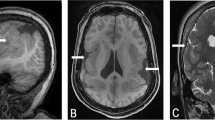
The patient was the first child of healthy parents without family history. The pregnancy was spontaneous. Gestational hypertension was diagnosed and treated from 14.5 weeks of gestation. Intrauterine growth retardation was initially diagnosed at 20 weeks. Ultrasound examination at 33.5 weeks demonstrated an increased heterogeneity of the placenta, with multiple hypoechogenic subchorial areas and confirmed growth retardation. At 36.5 weeks, high placental vascular resistance was also reported. Birth occurred at 38.5 weeks via Caesarean delivery, with birth weight 2,390 g, length 47.5 cm, and head circumference 32.5 cm. Placental weight was 260 g, with a mean diameter of 16 cm and mean thickness of 20 mm. Microscopic examination demonstrated a severe dystrophic pattern of the chorionic villi, with numerous terminal villi. Syncytiotrophoblast was by place retracted with clustered nuclei. There was no abnormal fibrin plaque. Anal stenosis, phimosis, and nonobstructive septal subaortic hypertrophy were observed soon after birth. Developmentally, the patient sat without support after the age of 9 months and walked at the age of 18 months. He was nonverbal until the age of 24 months. He was receiving speech and physical therapy. Dystonic attitudes of the neck with lateral inclination of the head was first reported at the age of 1 year. Jerky movements of the neck, right shoulder, and right arm appeared at 2 years. The jerks were seen in short, irregularly repeated periods. Dystonic attitude of the right arm was also present, largely accentuated by stress. The parents reported a decreased voluntary use of the right arm. The diagnosis of M-D syndrome was thereon proposed. Trihexyphenidyl associated with diazepam treatment started at the age of 2 years 4 months did not fully control dystonia. Several episodes of dystonic attitude occurring several times a month in the legs at night were also reported. Association of levodopa and benserazide started at age 4 accentuated the dystonia and was stopped. Spontaneous disappearance of jerky movements was reported at age 6. At the time of this report, at age 7 years, right arm dystonia was still present, with poor handwriting and more pronounced leg dystonia. Electroencephalogram (EEG) was unremarkable, and brain imaging, including scanner and magnetic resonance imaging (MRI), was normal. Physical examination revealed light skin color and a facial dysmorphism, including low eyebrows, anteverted nostrils, long philtrum, and thick lower lip (Fig. 1a).
a Facial appearance of the patient at 1 year of age. b Facial appearance of the patient at 5 years of age. Written consent to publish this photograph was obtained from the parents. c GTG-banded chromosomes obtained from lymphocytes of the proband: karyotype 46,XY,t(7;9)(q21;p23). d Fluorescent in situ hybridization (FISH) to metaphase using clone RP11-273O15 (934847754-94032828) encompassing SGCE and PEG10 genes showing one signal on normal chromosome 7 and absence of signal on der(7) (arrow). e FISH to metaphase using clone RP11-117J18 (12628464-12795950) showing one signal on normal chromosome 9 and absence of signal on der(9) (arrow)
Materials and methods
Cytogenetics and molecular cytogenetics
Blood samples from the patient and his parents were obtained after parental informed consent. Chromosome analysis was performed on peripheral blood lymphocytes by means of GTG banding. Fluorescence in-situ hybridization analysis (FISH) with whole-chromosome painting probes for chromosomes 7 and 9 (Vysis-Abbott, Des Plaines, IL, USA) was performed on the patient’s metaphases. Other FISH experiments were conducted with bacterial artificial chromosome (BAC) clones containing chromosomes 7- or 9-specific sequences from several locations (Table 1), in accordance with publicly available genome resources [National Center for Biotechnology Information (NCBI) Map Viewer : http://www.ncbi.nlm.nih.gov; Santa Cruz Human Genome Browser : http://www.genome.ucsc.edu]. BAC probes were obtained from the RPCI-1 and RPCI-11 libraries (BACPAC Resources Center, CHORI, Oakland, CA, USA) and selected according to their positions on chromosomes 7 or 9. BAC deoxyribonucleic acid (DNA) was labeled with biotin by nick translation. The labeled probes were visualized with fluorescein isothiocyanate–avidin (Vector Laboratories, Burlingame, CA, USA).
Molecular investigations
Genotyping of the patient and his parents was performed with ten chromosome 7 microsatellite markers: D7S657, D7S652, D7S2430, D7S1820, D7S3050, COL1A2, ESG, D7S2482, D7S2431, and D7S821, and three chromosome 9 microsatellite markers: D9S759, D9S268, and D9S254 selected from the Genome Database (http://www.gdb.org). Polymerase chain reaction (PCR) was performed using standard procedures with both locus specific primers. Each reaction contained one fluorescently labeled primer. DNA fragments were analyzed on an ABI Prism 310 Genetic Analyzer, and fragment sizes were determined with the GeneScan software (Applied Biosystems, Foster City, CA, USA).
On quantitative multiplex PCR of short fluorescent fragments (QMPSF) analysis, short exonic fragments of the 11 SGCE coding exons were simultaneously PCR amplified using dye-labeled primers corresponding to unique sequences. Two additional fragments, corresponding to exon 13 of the PBGD gene located on chromosome 11 and exon 8 of the NHS gene located on chromosome X, were coamplified as controls. PCR was performed in a final volume of 25 μl containing 100 ng of genomic DNA, 0.3–0.9 mM of each primer, 200 mM deoxynucleotide triphosphate (dNTP), 1.5 mM magnesium chloride (MgCl2), 10% of dimethylsulfoxide (DMSO), and 1U of Taq DNA polymerase (ABgene, Courtaboeuf, France). The PCR consisted of 23 cycles of 94°C for 10 s, 50°C for 15 s, and 72°C for 20 s, preceded by an initial denaturation step of 5 min at 94°C, and followed by a final extension of 5 min at 72°C. One microliter of the PCR product was resuspended in a mix containing 10 μl of deionized formamide and 0.25 μl of GeneScan-400 Rox (PE Applied Biosystems). After denaturation for 3 min at 90°C, 1 ml of each sample was loaded on an Applied Biosystems model 3100 automated sequencer. Data were analyzed using the GeneScan software. Electropherograms were superimposed onto those generated from a normal control DNA by adjusting to the same level the peaks obtained for the control amplicon, and the heights of the corresponding peaks were then compared between the different samples.
We used GC-tag-modified bisulfite genomic DNA sequencing, as described by Han et al. (2006), for simplified evaluation of DNA methylation sites on SGCE promoter. Genomic DNA was modified by EpiTect Bisulfite kit (Qiagen, France). The sens strand of bisulfite-modified genomic DNA was amplified with methylation-unspecific primers BSTx_F and BSTx_R specific for SGCE promoter, as described by Grabowski et al. (2003). The first-round PCR products were then used as template (1 μl) for the second-round PCR with tagged primers. PCR products were then purified with NucleoSpin® Extract II kit (Macherey-Nagel, SARL, Hoerdt, France) and subjected to sequencing using the tag-targeted sequencing primers.
Results
Cytogenetic and molecular cytogenetic investigations
GTG-banded chromosomes obtained from the proband’s lymphocytes revealed an apparently balanced translocation between chromosomes 7 and 9: 46,XY,t(7;9)(q21;p23) (Fig. 1b). Maternal and paternal karyotypes were normal. Whole-chromosome painting with chromosome 7 and chromosome 9 probes confirmed the translocation and excluded a more complex chromosome rearrangement involving other chromosomes (data not shown). FISH analysis with BAC probes showed interstitial cryptic deletions at the 7q21 and the 9p23 breakpoint regions (Table 1). The 7q21 deletion extended from clone RP11-258G9 (90443066-90628099) to clone RP1-133P16 (94070135-94135897). According to the University of California Santa Cruz (UCSC) genome browser (2004), these results revealed a loss spanning a 3.7-Mb genomic interval from 7q21.13 to 7q21.3 (Figs. 1c, 3a). An additional cryptic deletion of 5.2 Mb on chromosome 9p23 was identified from clone RP11-338L20 (8971678-9132717) to clone RP11-79B (914040094-14171677) (Figs. 1d, 3b).
Molecular studies
To establish the parent of origin of the deletions, we performed PCR with ten chromosome 7 microsatellite markers and with three chromosome 9 microsatellite markers. Three markers (D7S657, D7S1820, and D7S821) were informative for chromosome 7: D7S657 and D7S1820 revealed loss of the paternal allele (Fig. 2a), and D7S821 was outside the deleted region. D9S759 was informative for chromosome 9 and revealed loss of the paternal allele (Fig. 2b). The QMPSF profiles revealed a 50% reduction of peaks corresponding to the 11 SGCE amplicons (data not shown). Parents were tested by the same QMPSF analysis, which showed that they did not harbor the SGCE deletion.
a Paternal parent of origin of the deleted region on chromosome 7 using the D7S657 microsatellite marker. The patient inherited one 244-bp allele from his mother. b Paternal parent of origin of the deleted region on chromosome 9 using the D9S759 microsatellite marker. The patient inherited one 337-bp allele from his mother
We determined the methylation pattern of SGCE promoter for the patient, his father, his mother, and two normal controls. The amplified product of first-round PCR extended from position −773 to −1,148 and contained 25 CpG dinucleotides: from −48 to −72. Sequencing of father, mother, and normal controls showed both methylated and unmethylated DNA. Sequencing of the patient revealed that CpG dinucleotides were completely methylated (data not shown).
Discussion
Detailed FISH and molecular studies of this apparently balanced de novo reciprocal translocation led to the identification of cryptic deletions of paternal origin near the translocation breakpoints. The deleted regions contained at least 25 genes on chromosome 7 and seven genes on chromosome 9 (Fig. 3). These results allowed us to perform a genotype–phenotype analysis. A large imprinted gene cluster was previously identified on chromosome 7q21: SGCE and PEG10 (Ono et al. 2001) are paternally expressed and PPP1R9A (Nakabayashi et al. 2004) and GNGT1 are maternally expressed. SGCE alterations are implicated in M-D. Described alterations are truncating-point mutations or whole-exon deletions that cause a shift of the translational reading frame and introduce a premature termination codon. Maternal imprinting of SGCE has been found to explain reduced penetrance. Only paternally inherited heterozygous alterations of the SGCE gene cause M-D (Muller et al. 2002; Grabowski et al. 2003). In our case, M-D was due to complete deletion of the paternal allele of the SGCE gene. The first case of M-D due to complete paternal deletion of SGCE was published by DeBerardinis et al. (2003). Four other cases with M-D and complete deletion of SGCE were recently described (Asmus et al. 2007; Grünewald et al. 2008). PEG10 is strongly expressed in the placenta and is essential for embryo development (Ono et al. 2006). Prenatal growth retardation observed in the proband may be due to complete deletion of the paternal allele of the PEG10 gene. Other genes in the deleted region are not imprinted; however, a phenotype can be due to haploinsufficiency of these genes. KRIT1 is responsible for 40% or more of familial cases of cerebral cavernous malformations (CCM). Haploinsufficiency of KRIT1 is the major mutational mechanism in CCM (Marini et al. 2004). Our patient showed no symptom of CCM, and cranial MRI was normal. Two patients with complete heterozygous deletion of KRIT1 and CCM were described. The first one harbored a single cavernous lesion in the left frontal lobe (Tzschach et al. 2007). The second presented several small lesions (Asmus et al. 2007). Both were asymptomatic. The absence of cavernous malformations in our case is probably due to the late onset of this disorder and the incomplete penetrance. The detection of a KRIT1 deletion therefore has pivotal consequences for a follow-up of neuroimaging with cranial MRI.
a Map of deleted genes, bacterial artificial chromosome (BAC) clones studied, and polymorphic markers in the 7q21 region. b Map of deleted genes, BAC clones studied, and polymorphic markers in the 9p23 region (from Database of Genomic Variants http://projects.tcag.ca/variation/)
The deletion region also contains the COL1A2 gene, which encodes the alpha-2 chain of type I collagen. In severe forms of osteogenesis imperfecta, COL1A2 mutations exert a dominant negative effect. However, COL1A2 haploinsufficiency could be implicated in very mild forms of osteogenesis imperfecta type I. Indeed, three patients have been previously reported (Asmus et al. 2007; Grünewald et al. 2008) with COL1A2 deletion and delayed skeletal development, osteoporosis, or joint problems. These data should guide the clinical follow-up to detect signs of bone and joint disease. ERVWE1 encodes syncitin and is strongly expressed in the placental syncytiotrophoblast (Mi et al. 2000). In pre-eclampsia syncytin, expression levels are dramatically reduced (Kudaka et al. 2008). Dysregulated syncytin expression leads to abnormalities in syncytiotrophoblast growth and chorionic villous growth and maturation, contributing to placental abnormalities seen in pre-eclampsia. In our case, gestational hypertension was diagnosed at 14.5 weeks and may have been due to ERVWE1 haploinsufficiency.
Nine other patients with an interstitial deletion in 7q21 and delineation of the breakpoint regions are described in the literature. Five out of ten deletions include the five genes we described earlier: SGCE, PEG10, KRIT1, COL1A2, and ERVWE1 (Fig. 4a). Four patients seemed to have the same distal breakpoint telomeric to PEG10 (patients 1, 3, 7, and 8). This breakpoint is located in a repetitive sequence of long and short interspersed nuclear elements (Asmus et al. 2007). Table 2 reviews phenotypes of these ten cases. The parent of origin of the deletion was determined in seven cases, and the deleted allele was always paternal. This might be explained by an increased sensitivity of meiotic and postmeiotic stages of spermatogenesis to the induction of large genomic deletions and translocations. De Gregori et al. (2007) analyzed the parent of origin of five deletions associated with translocations and 11 associated with complex chromosome rearrangements (CCR), and the deleted allele was always paternal. Thomas et al. (2006) showed that 84% of interstitial deletions of 35 studied were paternal. Intrauterine growth retardation, short stature, microcephaly, developmental delay, and facial dysmorphism are the more recurrent features. M-D was diagnosed in only six of the seven patients with paternal SGCE deletion. Age of onset of M-D can be in adolescence, and some patients may have not yet developed the M-D phenotype. The only two patients with KRIT1 deletion, at the age of 40 years, harbor CCM. Patients 2, 7, 8, and 10 present a very mild form of osteogenesis imperfecta type I due to COL1A2 haploinsufficiency. A significant hearing loss is observed in the four patients with SHFM1 deletion.
When comparing clinical signs of patients with paternal 7q21 deletion with those of Silver-Russell syndrome (SRS) with maternal uniparental disomy of the chromosome 7, a significant clinical overlap is observed. It suggests an involvement of paternally expressed imprinted gene on chromosome 7q21 in clinical phenotype. According to paternal expression of SGCE, SRS patients with matUPD(7) should be susceptible to develop M-D. The fact that no case is described may be due to the young age of described children with matUPD(7) and the difficulties for physicians to identify M-D. PEG10 (paternally expressed 10) could be involved in intrauterine growth delay observed in SRS with matUPD(7).
The deleted region on chromosome 9 overlaps with the candidate region for monosomy 9p syndrome (Kawara et al. 2006; Faas et al. 2007). Three genes, TYRP1, MPDZ, and NFIB included in the candidate region were deleted in our patient (Fig. 4b). Homozygous and compound heterozygous mutations in TYRP1 result in Rufous oculocutaneous albinism. TYRP1 may have caused light skin color in our patient. Haploinsufficiency at the NFIB locus in mice is involved in callosal agenesis and delayed lung maturation (Steele-Perkins et al. 2005). Kawara et al. (2006) reported a case of monosomy 9p and agenesis of the corpus callosum, which may be due to NFIB haploinsufficiency. Our patient shows dysmorphic features compatible with monosomy 9p syndrome: anteverted nostrils and long philtrum. Mental retardation and trigonocephaly, which are major features in monosomy 9p syndrome, are due to, respectively, ZDHHC21 and CER1 genes, which were not included in the deleted region in our case. In sum, we have shown deleted genes to be contributory to the patient’s phenotype, and our data support the hypothesis that apparently balanced chromosomal rearrangements are valuable biological landmarks for genes important in human development.
References
Asmus F, Hjermind LE, Dupont E, Wagenstaller J, Haberlandt E, Munz M, Strom TM, Gasser T (2007) Genomic deletion size at the epsilon-sarcoglycan locus determines the clinical phenotype. Brain 130:2736–2745
Courtens W, Vermeulen S, Wuyts W, Messiaen L, Wauters J, Nuytinck L, Peeters N, Storm K, Speleman F, Nothen MM (2005) An interstitial deletion of chromosome 7 at band q21: a case report and review. Am J Med Genet A 134:12–23
DeBerardinis RJ, Conforto D, Russell K, Kaplan J, Kollros PR, Zackai EH, Emanuel BS (2003) Myoclonus in a patient with a deletion of the epsilon-sarcoglycan locus on chromosome 7q21. Am J Med Genet A 121:31–36
De Gregori M, Ciccone R, Magini P, Pramparo T, Gimelli S, Messa J, Novara F, Vetro A, Rossi E, Maraschio P, Bonaglia MC, Anichini C, Ferrero GB, Silengo M, Fazzi E, Zatterale A, Fischetto R, Previdere C, Belli S, Turci A, Calabrese G, Bernardi F, Meneghelli E, Riegel M, Rocchi M, Guerneri S, Lalatta F, Zelante L, Romano C, Fichera M, Mattina T, Arrigo G, Zollino M, Giglio S, Lonardo F, Bonfante A, Ferlini A, Cifuentes F, Van Esch H, Backx L, Schinzel A, Vermeesch JR, Zuffardi O (2007) Cryptic deletions are a common finding in “balanced” reciprocal and complex chromosome rearrangements: a study of 59 cases. J Med Genet 44:750–752
Faas BH, de Leeuw N, Mieloo H, Bruinenberg J, de Vries BB (2007) Further refinement of the candidate region for monosomy 9p syndrome. Am J Med Genet A 143:2353–2356
Fukushima K, Nagai K, Tsukada H, Sugata A, Sugata K, Kasai N, Kibayashi N, Maeda Y, Gunduz M, Nishizaki K (2003) Deletion mapping of split hand/split foot malformation with hearing impairment: a case report. Int J Pediatr Otorhinolaryngol 67:1127–1132
Grabowski M, Zimprich A, Lorenz-Depiereux B, Kalscheuer V, Asmus F, Gasser T, Meitinger T, Strom TM (2003) The epsilon-sarcoglycan gene (SGCE), mutated in M-D syndrome, is maternally imprinted. Eur J Hum Genet 11:138–144
Grünewald A, Djarmati A, Lohmann-Hedrich K, Farrell K, Zeller JA, Allert N, Papengut F, Petersen B, Fung V, Sue CM, O’Sullivan D, Mahant N, Kupsch A, Chuang RS, Wiegers K, Pawlack H, Hagenah J, Ozelius LJ, Stephani U, Schuit R, Lang AE, Volkmann J, Münchau A, Klein C (2008) Myoclonus-dystonia: significance of large SGCE deletions. Hum Mutat 29:331–332
Haberlandt E, Loffler J, Hirst-Stadlmann A, Stockl B, Judmaier W, Fischer H, Heinz-Erian P, Muller T, Utermann G, Smith RJ, Janecke AR (2001) Split hand/split foot malformation associated with sensorineural deafness, inner and middle ear malformation, hypodontia, congenital vertical talus, and deletion of eight microsatellite markers in 7q21.1–q21.3. J Med Genet 38:405–409
Han W, Cauchi S, Herman JG, Spivack SD (2006) DNA methylation mapping by tag-modified bisulfite genomic sequencing. Anal Biochem 355:50–61
Kawara H, Yamamoto T, Harada N, Yoshiura K, Niikawa N, Nishimura A, Mizuguchi T, Matsumoto N (2006) Narrowing candidate region for monosomy 9p syndrome to a 4.7-Mb segment at 9p22.2–p23. Am J Med Genet A 140:373–377
Kudaka W, Oda T, Jinno Y, Yoshimi N, Aoki Y (2008) Cellular localization of placenta-specific human endogenous retrovirus (HERV) transcripts and their possible implication in pregnancy-induced hypertension. Placenta 29:282–289
Marini V, Ferrera L, Pigatto F, Origone P, Garre C, Dorcaratto A, Viale G, Alberti F, Mareni C (2004) Search for loss of heterozygosity and mutation analysis of KRIT1 gene in CCM patients. Am J Med Genet A 130:98–101
Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC Jr, McCoy JM (2000) Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789
Muller B, Hedrich K, Kock N, Dragasevic N, Svetel M, Garrels J, Landt O, Nitschke M, Pramstaller PP, Reik W, Schwinger E, Sperner J, Ozelius L, Kostic V, Klein C (2002) Evidence that paternal expression of the epsilon-sarcoglycan gene accounts for reduced penetrance in M-D. Am J Hum Genet 71:1303–1311
Nakabayashi K, Makino S, Minagawa S, Smith AC, Bamforth JS, Stanier P, Preece M, Parker-Katiraee L, Paton T, Oshimura M, Mill P, Yoshikawa Y, Hui CC, Monk D, Moore GE, Scherer SW (2004) Genomic imprinting of PPP1R9A encoding neurabin I in skeletal muscle and extra-embryonic tissues. J Med Genet 41:601–608
Nardocci N, Zorzi G, Barzaghi C, Zibordi F, Ciano C, Ghezzi D, Garavaglia B (2008) Myoclonus-dystonia syndrome: clinical presentation, disease course, and genetic features in 11 families. Mov Disord 23:28–34
Ono R, Kobayashi S, Wagatsuma H, Aisaka K, Kohda T, Kaneko-Ishino T, Ishino F (2001) A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73:232–237
Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, Hino T, Suzuki-Migishima R, Ogonuki N, Miki H, Kohda T, Ogura A, Yokoyama M, Kaneko-Ishino T, Ishino F (2006) Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet 38:101–106
Steele-Perkins G, Plachez C, Butz KG, Yang G, Bachurski CJ, Kinsman SL, Litwack ED, Richards LJ, Gronostajski RM (2005) The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol 25:685–698
The University of California Santa Cruz (UCSC) genome browser (2004) UCSC Genome Bioinformatics Group, Santa Cruz. http://genome.ucsc.edu Accessed May 2004
Thomas NS, Durkie M, Van Zyl B, Sanford R, Potts G, Youings S, Dennis N, Jacobs P (2006) Parental and chromosomal origin of unbalanced de novo structural chromosome abnormalities in man. Hum Genet 119:444–450
Tzschach A, Menzel C, Erdogan F, Schubert M, Hoeltzenbein M, Barbi G, Petzenhauser C, Ropers HH, Ullmann R, Kalscheuer V (2007) Characterization of a 16 Mb interstitial chromosome 7q21 deletion by tiling path array CGH. Am J Med Genet A 143:333–337
Zimprich A, Grabowski M, Asmus F, Naumann M, Berg D, Bertram M, Scheidtmann K, Kern P, Winkelmann J, Muller-Myhsok B, Riedel L, Bauer M, Muller T, Castro M, Meitinger T, Strom TM, Gasser T (2001) Mutations in the gene encoding epsilon-sarcoglycan cause M-D syndrome. Nat Genet 29:66–69
Acknowledgments
We express our sincere gratitude to the patient and his family. We thank the cytogenetics and molecular genetics staff at the Nancy University Hospital for their expert technical assistance. This study was supported by grants from Ministère de la Recherche (EA 4002).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bonnet, C., Grégoire, MJ., Vibert, M. et al. Cryptic 7q21 and 9p23 deletions in a patient with apparently balanced de novo reciprocal translocation t(7;9)(q21;p23) associated with a dystonia-plus syndrome: paternal deletion of the epsilon-sarcoglycan (SGCE) gene. J Hum Genet 53, 876–885 (2008). https://doi.org/10.1007/s10038-008-0321-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-008-0321-z