Abstract
As a key step in achieving low-cost, easily accessible anti-cancer therapy for low- and middle-income countries, we recently established the scientific basis for the folkloric use of Artocarpus altilis for the treatment of cancer by investigating the geranyl dihydrochalcone (CG-901) content and its interference with signal transducer and activator of transcription 3 (STAT3) phosphorylation and blockage of further downstream signaling. In the current study, the CG-901 upstream target was queried by chemical fingerprinting similarity assessment, semi-empirical (PM6ESCF) QMMM and molecular dynamics (MD) simulation. Moderate (∼0.4) to high (∼0.7) Tanimoto scores were found when the CG-901 scaffold was compared to ligands co-crystallized with Janus kinases (JAK) 1–3. High negative energy values were obtained when the CG-901 was treated semi-empirically (PM6ESCF) within the classical field of JAK (1–3). Multiple nanosecond MD simulations showed that CG-901 did not cause any large structural perturbations in the nucleotide-binding, activation and catalytic loops within the kinase (JH1) domain of JAK (1–3); however, it reduced the energy required to attain metastability along the path to energy minima conformation. In comparison to JAK1 and Apo-state JAK2, JAK2-bound CG-901 exhibited a highly re-organized key intra-domain protein network; indicating atomic level interference with inter-residue communication. In conclusion, CG-901 isolated from A. altilis represents a broad-spectrum JAK inhibitor, which may underlie the mechanism of STAT3 phosphorylation blockage.

Upper panel Janus kinase 2 upstream signaling pathway. Lower panel Apo-JAK2 (left) and CG-901-bound JAK2 (right)




Similar content being viewed by others
References
Andre N, Banavali S, Snihur Y, Pasquier E (2013) Has the time come for metronomics in low-income and middle-income countries? The Lancet. Oncology 14:e239–e248
Luqmani YA (2005) Mechanisms of drug resistance in cancer chemotherapy. Med Princ Pract: Int J Kuwait Univ Health Sci Centre 14(Suppl 1):35–48
Al-Lazikani B, Banerji U, Workman P (2012) Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol 30:679–692
Stritzker J, Szalay AA (2013) Single-agent combinatorial cancer therapy. Proc Natl Acad Sci USA 110:8325–8326
Chang-Yew Leow C, Gerondakis S, Spencer A (2013) MEK inhibitors as a chemotherapeutic intervention in multiple myeloma. Blood Cancer J 3, e105
Hamburg MA, Collins FS (2010) The path to personalized medicine. N Engl J Med 363:301–304
Jeon YJ, Jung SN, Chang H, Yun J, Lee CW, Lee J, Choi S, Nash O, Han DC, Kwon BM (2015)Artocarpus altilis (Parkinson) Fosberg extracts and geranyl dihydrochalcone inhibit STAT3 activity in prostate cancer DU145 cells. Phytother Res 29:749–756
Tulunay A, Dozmorov MG, Ture-Ozdemir F, Yilmaz V, Eksioglu-Demiralp E, Alibaz-Oner F, Ozen G, Wren JD, Saruhan-Direskeneli G, Sawalha AH, Direskeneli H (2015) Activation of the JAK/STAT pathway in Behcet’s disease. Genes Immun 16:176
Pinz S, Unser S, Brueggemann S, Besl E, Al-Rifai N, Petkes H, Amslinger S, Rascle A (2014) The synthetic alpha-bromo-2′,3,4,4′-tetramethoxychalcone (alpha-Br-TMC) inhibits the JAK/STAT signaling pathway. PLoS One 9, e90275
Looyenga BD, Hutchings D, Cherni I, Kingsley C, Weiss GJ, Mackeigan JP (2012) STAT3 is activated by JAK2 independent of key oncogenic driver mutations in non-small cell lung carcinoma. PLoS One 7, e30820
Bernstein FC, Koetzle TF, Williams GJ, Meyer EF Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M (1977) The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol 112:535–542
Hurley CA, Blair WS, Bull RJ, Chang C, Crackett PH, Deshmukh G, Dyke HJ, Fong R, Ghilardi N, Gibbons P, Hewitt PR, Johnson A, Johnson T, Kenny JR, Kohli PB, Kulagowski JJ, Liimatta M, Lupardus PJ, Maxey RJ, Mendonca R, Narukulla R, Pulk R, Ubhayakar S, van Abbema A, Ward SI, Waszkowycz B, Zak M (2013) Novel triazolo-pyrrolopyridines as inhibitors of Janus kinase 1. Bioorg Med Chem Lett 23:3592–3598
Brasca MG, Gnocchi P, Nesi M, Amboldi N, Avanzi N, Bertrand J, Bindi S, Canevari G, Casero D, Ciomei M, Colombo N, Cribioli S, Fachin G, Felder ER, Galvani A, Isacchi A, Motto I, Panzeri A, Donati D (2015) Novel pyrrole carboxamide inhibitors of JAK2 as potential treatment of myeloproliferative disorders. Bioorg Med Chem 23:2387–2407
Duan JJ, Lu Z, Jiang B, Yang BV, Doweyko LM, Nirschl DS, Haque LE, Lin S, Brown G, Hynes J Jr, Tokarski JS, Sack JS, Khan J, Lippy JS, Zhang RF, Pitt S, Shen G, Pitts WJ, Carter PH, Barrish JC, Nadler SG, Salter-Cid LM, McKinnon M, Fura A, Schieven GL, Wrobleski ST (2014) Discovery of pyrrolo[1,2-b]pyridazine-3-carboxamides as Janus kinase (JAK) inhibitors. Bioorg Med Chem Lett 24:5721–5726
Molecular Operating Environment (MOE) (2015) Chemical Computing Group, 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7
Heifets A, Lilien RH (2010) LigAlign: flexible ligand-based active site alignment and analysis. J Mol Graph Model 29:93–101
Jo S, Kim T, Iyer VG, Im W (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 29:1859–1865
Florová P, Sklenovsky P, Banáš P, Otyepka M (2010) Explicit water models affect the specific solvation and dynamics of unfolded peptides while the conformational behavior and flexibility of folded peptides remain intact. J Chem Theory Comput 6:3569–3579
Gotz AW, Clark MA, Walker RC (2014) An extensible interface for QM/MM molecular dynamics simulations with AMBER. J Comput Chem 35:95–108
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718
Huang J, MacKerell AD Jr (2013) CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J Comput Chem 34:2135–2145
Berendsen HJCP, van Gunsteren JPM, DiNola WF, Haak AJR (1984) Molecular-dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N log (N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Van Gunsteren WF, Berendsen HJC (1988) A leap-frog algorithm for stochastic dynamics. Mol Simul 1:173–185
Seeliger D, de Groot BL (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des 24:417–422
Halgren TA (1996) Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 17:490–519
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Wolfram Research (2014) MATHEMATICA. vol. version 10.0. Wolfram Research, Champaign, IL
Li T, Froeyen M, Herdewijn P (2010) Insight into ligand selectivity in HCV NS5B polymerase: molecular dynamics simulations, free energy decomposition and docking. J Mol Model 16:49–59
Omotuyi OI, Nagai J, Ueda H (2015) Lys39-lysophosphatidate carbonyl oxygen interaction locks LPA1 N-terminal cap to the orthosteric Site and partners Arg124 during receptor activation. Sci Rep 5:13343
Sethi A, Eargle J, Black AA, Luthey-Schulten Z (2009) Dynamical networks in tRNA: protein complexes. Proc Natl Acad Sci USA 106:6620–6625
Koukos PI, Glykos NM (2013) Grcarma: a fully automated task-oriented interface for the analysis of molecular dynamics trajectories. J Comput Chem 34:2310–2312
Jatiani SS, Cosenza SC, Reddy MV, Ha JH, Baker SJ, Samanta AK, Olnes MJ, Pfannes L, Sloand EM, Arlinghaus RB, Reddy EP (2010) A non-ATP-competitive dual inhibitor of JAK2 and BCR-ABL kinases: elucidation of a novel therapeutic spectrum based on substrate competitive inhibition. Genes Cancer 1:331–345
Antonysamy S, Hirst G, Park F, Sprengeler P, Stappenbeck F, Steensma R, Wilson M, Wong M (2009) Fragment-based discovery of JAK-2 inhibitors. Bioorg Med Chem Lett 19:279–282
Duan J, Dixon SL, Lowrie JF, Sherman W (2010) Analysis and comparison of 2D fingerprints: insights into database screening performance using eight fingerprint methods. J Mol Graph Model 29:157–170
Wang T, Duffy JP, Wang J, Halas S, Salituro FG, Pierce AC, Zuccola HJ, Black JR, Hogan JK, Jepson S, Shlyakter D, Mahajan S, Gu Y, Hoock T, Wood M, Furey BF, Frantz JD, Dauffenbach LM, Germann UA, Fan B, Namchuk M, Bennani YL, Ledeboer MW (2009) Janus kinase 2 inhibitors. Synth Charact Nov Polycycl Azaindole J Med Chem 52:7938–7941
Rathore RS, Sumakanth M, Reddy MS, Reddanna P, Rao AA, Erion MD, Reddy MR (2013) Advances in binding free energies calculations: QM/MM-based free energy perturbation method for drug design. Curr Pharm Des 19:4674–4686
Leonard WJ, O’Shea JJ (1998) Jaks and STATs: biological implications. Annu Rev Immunol 16:293–322
Yoshikawa H, Matsubara K, Qian GS, Jackson P, Groopman JD, Manning JE, Harris CC, Herman JG (2001) SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat Genet 28:29–35
Feng J, Witthuhn BA, Matsuda T, Kohlhuber F, Kerr IM, Ihle JN (1997) Activation of Jak2 catalytic activity requires phosphorylation of Y1007 in the kinase activation loop. Mol Cell Biol 17:2497–2501
Lucet IS, Fantino E, Styles M, Bamert R, Patel O, Broughton SE, Walter M, Burns CJ, Treutlein H, Wilks AF, Rossjohn J (2006) The structural basis of Janus kinase 2 inhibition by a potent and specific pan-Janus kinase inhibitor. Blood 107:176–183
Huse M, Kuriyan J (2002) The conformational plasticity of protein kinases. Cell 109:275–282
Alicea-Velazquez NL, Boggon TJ (2011) The use of structural biology in Janus kinase targeted drug discovery. Curr Drug Targets 12:546–555
West K (2009) CP-690550, a JAK3 inhibitor as an immunosuppressant for the treatment of rheumatoid arthritis, transplant rejection, psoriasis and other immune-mediated disorders. Curr Opin Investig Drugs 10:491–504
Lin Q, Lai R, Chirieac LR, Li C, Thomazy VA, Grammatikakis I, Rassidakis GZ, Zhang W, Fujio Y, Kunisada K, Hamilton SR, Amin HM (2005) Constitutive activation of JAK3/STAT3 in colon carcinoma tumors and cell lines: inhibition of JAK3/STAT3 signaling induces apoptosis and cell cycle arrest of colon carcinoma cells. Am J Pathol 167:969–980
Kumar N, Mishra J, Narang VS, Waters CM (2007) Janus kinase 3 regulates interleukin 2-induced mucosal wound repair through tyrosine phosphorylation of villin. J Biol Chem 282:30341–30345
O’Shea JJ, Gadina M, Schreiber RD (2002) Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell 109(Suppl):S121–S131
Park ES, Kim H, Suh JM, Park SJ, You SH, Chung HK, Lee KW, Kwon OY, Cho BY, Kim YK, Ro HK, Chung J, Shong M (2000) Involvement of JAK/STAT (Janus kinase/signal transducer and activator of transcription) in the thyrotropin signaling pathway. Mol Endocrinol 14:662–670
Chen YW, Guo T, Shen L, Wong KY, Tao Q, Choi WW, Au-Yeung RK, Chan YP, Wong ML, Tang JC, Liu WP, Li GD, Shimizu N, Loong F, Tse E, Kwong YL, Srivastava G (2015) Receptor-type tyrosine-protein phosphatase kappa directly targets STAT3 activation for tumor suppression in nasal NK/T-cell lymphoma. Blood 125:1589–1600
Acknowledgment
This work was partly supported by the H3Africa Bioinformatics Network (H3ABioNet) grant funded by NIH Common Fund Award/NHGRI (Grant Number U41HG006941).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(MOV 8.61 mb)
Supplementary Fig. S1
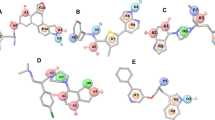
Binding signature and dynamics of JAK3-JH1 CG-901 or 3QX bound states. a Free energy surface plots of Apo-JAK3 (I), JAK3-QUP (ii), and JAK3-CG-901 complex. b Upper plane) Representative low-energy conformation of 3QX (red stick)-JAK3 (blue cartoon) complex, green stick JAK3 residues interacting with 3QX (cyan stick). Lower plane) Representative low-energy conformation of CG-901 (green stick)-JAK3 (blue cartoon) complex; cyan stick represents the JAK3 residues interacting with CG-901 (green stick). c Line plots of χ1 (I, upper plane) and χ2 (I, lower plane) dihedral angle distribution of Tyr1007 along the trajectory, (ii upper plane) root mean square deviation of the nucleotide binding loop, (ii lower plane) catalytic loop, (iii, upper plane) activation loop during the trajectories. (iii, lower plane) center of mass distance between tyrosine 1007 and aspartate 1004 during the course of the simulation. d Cartoon representation of apo-JAK3 (I), 3QX-bound (ii), and CG-901-bound (iii) JAK3. Blue spheres and line (thickness represents weight) represent critical network. Lines plot are color coded: Red Apo-protein, Blue lines 3QX-bound JAK3, green lines represent CG-901-bound JAK3. (PDF 5.70 mb)
Rights and permissions
About this article
Cite this article
Nash, O., Omotuyi, O., Lee, J. et al. Artocarpus altilis CG-901 alters critical nodes in the JH1-kinase domain of Janus kinase 2 affecting upstream JAK/STAT3 signaling. J Mol Model 21, 280 (2015). https://doi.org/10.1007/s00894-015-2821-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-015-2821-z