Abstract
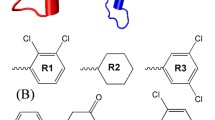
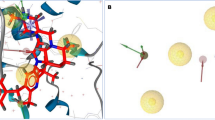
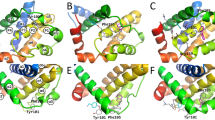
Protein-protein interactions are abundant in signal transduction pathways and thus of crucial importance in the regulation of apoptosis. However, designing small-molecule inhibitors for these potential drug targets is very challenging as such proteins often lack well-defined binding pockets. An example for such an interaction is the binding of the anti-apoptotic BIR2 domain of XIAP to the pro-apoptotic caspase-3 that results in the survival of damaged cells. Although small-molecule inhibitors of this interaction have been identified, their exact binding sites on XIAP are not known as its crystal structures reveal no suitable pockets. Here, we apply our previously developed protocol for identifying transient binding pockets to XIAP-BIR2. Transient pockets were identified in snapshots taken during four different molecular dynamics simulations that started from the caspase-3:BIR2 complex or from the unbound BIR2 structure and used water or methanol as solvent. Clustering of these pockets revealed that surprisingly many pockets opened in the flexible linker region that is involved in caspase-3 binding. We docked three known inhibitors into these transient pockets and so determined five putative binding sites. In addition, by docking two inactive compounds of the same series, we show that this protocol is also able to distinguish between binders and nonbinders which was not possible when docking to the crystal structures. These findings represent a first step toward the understanding of the binding of small-molecule XIAP-BIR2 inhibitors on a molecular level and further highlight the importance of considering protein flexibility when designing small-molecule protein-protein interaction inhibitors.




Similar content being viewed by others
References
Wells AL, McClendon CL (2007) Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 450:1001–1009
Roy N, Deveraux QL, Takahashi R, Salvesen GS, Reed JC (1997) The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J 16:6914–6925
Deveraux QL, Roy N, Stennicke HR, Van Arsdale T, Zhou Q, Srinivasula SM, Alnemri ES, Salvesen GS, Reed JC (1998) IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J 17:2215–2223
Reed JC (2001) The survivin saga goes in vivo. J Clin Invest 108:965–969
Cohen GM (1997) Caspases: the executioners of apoptosis. Biochem J 326:1–16
Thornberry NA, Lazebnik Y (1998) Caspases: enemies within. Science 281:1312–1316
Tamm I, Kornblau SM, Segall H, Krajewski S, Welsh K, Kitada S, Scudiero DA, Tudor G, Qui YH, Monks A, Andreeff M, Reed JC (2000) Expression and prognostic significance of IAP-family genes in human cancers and myeloid leukemias. Clin Cancer Res 6:1796–1803
Takahashi R, Deveraux Q, Tamm I, Welsh K, Assa-Munt N, Salvesen GS, Reed JC (1998) A single BIR domain of XIAP sufficient for inhibiting caspases. J Biol Chem 14:7787–7790
Deveraux QL, Leo E, Stennicke HR, Welsh K, Salvesen GS, Reed JC (1999) Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases. EMBO J 18:5242–5251
Du C, Fang M, Li Y, Li L, Wang X (2000) Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102:33–42
Riedl SJ, Renatus M, Schwarzenbacher R, Zhou Q, Sun C, Fesik SW, Liddington RC, Salvesen GS (2001) Structural basis for the inhibition of caspase-3 by XIAP. Cell 104:791–800
Chai J, Shiozaki E, Srinivasula SM, Wu Q, Datta P, Alnemri ES, Shi Y (2001) Structural basis of caspase-7 inhibition by XIAP. Cell 104:769–780
Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y (2003) Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell 11:519–527
Liu Z, Sun C, Olejniczak ET, Meadows RP, Betz SF, Oost T, Herrmann J, Wu JC, Fesik SW (2000) Structural basis for binding of Smac/Diablo to the XIAP BIR3 domain. Nature 408:1004–1008
Wu G, Chai J, Suber TL, Wu JW, Du C, Wang X, Shi Y (2000) Structural basis of IAP recognition by Smac/Diablo. Nature 408:1008–1012
Wist AD, Gu L, Riedl SJ, Shi Y, McLendon GL (2007) Structure-activity based study of the Smac-binding pocket within the BIR3 domain of XIAP. Bioorg Med Chem 15:2935–2943
Sun H, Stuckey JA, Nikolovska-Coleska Z, Qin D, Meagher JL, Qiu S, Lu J, Yang CY, Saito NG, Wang S (2008) Structure-based design, synthesis, evaluation, and crystallographic studies of conformationally constrained Smac mimetics as inhibitors of the X-linked inhibitor of apoptosis protein (XIAP). J Med Chem 51:7169–7180
Mastrangelo E, Cossu F, Milani M, Sorrentino G, Lecis D, Delia D, Manzoni L, Drago C, Seneci P, Scolastico C, Rizzo V, Bolognesi M (2008) Targeting the X-linked inhibitor of apoptosis protein through 4-substituted azabicyclo[5.3.0]alkane Smac mimetics. Structure, activity, and recognition principles. J Mol Biol 384:673–689
Nikolovska-Coleska Z, Meagher JL, Jiang S, Yang CY, Qiu S, Roller PP, Stuckey JA, Wang S (2008) Interaction of a cyclic, bivalent Smac mimetic with the x-linked inhibitor of apoptosis protein. Biochemistry 47:9811–9824
Cossu F, Milani M, Mastrangelo E, Vachette P, Servida F, Lecis D, Canevari G, Delia D, Drago C, Rizzo V, Manzoni L, Seneci P, Scolastico C, Bolognesi M (2009) Structural basis for bivalent Smac-mimetics recognition in the IAP protein family. J Mol Biol 392:630–644
Schimmer AD, Welsh K, Pinilla C, Wang Z, Krajewska M, Bonneau MJ, Pedersen IM, Kitada S, Scott FL, Bailly-Maitre B, Glinsky G, Scudiero D, Sausville E, Salvesen G, Nefzi A, Ostresh JM, Houghten RA, Reed JC (2004) Small-molecule antagonists of apoptosis suppressor XIAP exhibit broad antitumor activity. Cancer Cell 5:25–35
Wang Z, Cuddy M, Samuel T, Welsh K, Schimmer A, Hanaii F, Houghten R, Pinilla C, Reed JC (2004) Cellular, biochemical, and genetic analysis of mechanism of small molecule IAP inhibitors. J Biol Chem 279:48168–48176
Kater AP, Dicker F, Mangiola M, Welsh K, Houghten R, Ostresh J, Nefzi A, Reed JC, Pinilla C, Kipps TJ (2005) Inhibitors of XIAP sensitize CD40-activated chronic lymphocytic leukemia cells to CD95-mediated apoptosis. Blood 106:1742–1748
Scott FL, Denault JB, Riedl SJ, Shin H, Renatus M, Salvesen GS (2005) XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved mechanism of IAPs. EMBO J 24:645–655
Sun C, Cai M, Gunasekera AH, Meadows RP, Wang H, Chen J, Zhang H, Wu W, Xu N, Ng SC, Fesik SW (1999) NMR structure and mutagenesis of the inhibitor-of-apoptosis protein XIAP. Nature 401:818–822
Eyrisch S, Helms V (2007) Transient pockets on protein surfaces involved in protein-protein interaction. J Med Chem 50:3457–3464
Eyrisch S, Helms V (2009) What induces pocket openings on protein surface patches involved in protein-protein interactions? J Comput Aid Mol Des 23:73–86
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19:1639–1662
Berwanger A, Eyrisch S, Schuster I, Helms V, Bernhardt R (2010) Polyamines: naturally occurring small molecule modulators of electrostatic protein-protein interactions. J Inorg Biochem 104:118–125
Lerner MG, Bowman AL, Carlson HA (2007) Incorporating dynamics in E. coli dihydrofolate reductase enhances structure-based drug discovery. J Chem Inf Model 47:2358–2365
Bowman AL, Lerner MG, Carlson HA (2007) Protein flexibility and species specificity in structure-based drug discovery: dihydrofolate reductase as a test system. J Am Chem Soc 129:3634–3640
Bogan AA, Thorn KS (1998) Anatomy of hot spots in protein interfaces. J Mol Biol 280:1
Clackson T, Wells J (1995) A hot-spot of binding energy in a hormone-receptor interface. Science 267:383–386
Moreira IS, Fernandes PA, Ramos MJ (2007) Hot spots-a review of the protein-protein interface determinant amino-acid residues. Proteins 68:803–812
Halperin I, Wolfson H, Nussinov R (2004) Protein-protein interactions: coupling of structurally conserved residues and of hot spots across interfaces. Implications for docking. Structure 12:1027–1038
Keskin O, Ma B, Nussinov R (2005) Hot regions in protein-protein interactions: the organization and contribution of structurally conserved hot spot residues. J Mol Biol 345:1281–1294
Kortemme T, Baker D (2002) A simple physical model for binding energy hot spots in protein-protein complexes. Proc Natl Acad Sci USA 99:14116–14121
Guerois R, Nielsen J, Serrano L (2002) Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol 320:369–387
Tuncbag N, Gursoy A, Keskin O (2009) Identification of computational hot spots in protein interfaces: combining solvent accessibility and inter-residue potentials improves the accuracy. Bioinformatics 25:1513–1520
Bylaska EJ, de Jong WA, Kowalski K, Straatsma TP, Valiev M et al (2006) NWChem, a computational chemistry package for parallel computers, version 5.0, Pacific Northwest National Laboratory, Richland, Washington 99352-0999, USA
Becke AD (1997) Density-functional thermochemistry. V. Systematic optimization of exchange-correlation functionals. J Chem Phys 107:8554
Bayly CI, Cieplak P, Cornell W, Kollman PA (1993) A well-behaved electrostatic potential based method using charge restraints for determining atom-centered charges: the RESP model. J Phys Chem 97:10269–10280
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the opls all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236
Merz KM (1991) Carbon dioxide binding to human carbonic anhydrase ii. J Am Chem Soc 113:406–411
Lindahl E, Hess B, van der Spoel D (2001) GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Model 7:306–317
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Darden T, York D, Pedersen L (1993) Particle mesh ewald: an n log(n) method for ewald sums in large systems. J Chem Phys 98:10089–10092
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Brady GP Jr, Stouten PF (2000) Fast prediction and visualization of protein binding pockets with pass. J Comput Aided Mol Des 14:383–401
Gasteiger J, Marsili M (1980) Iterative partial equilibration of orbital electronegativity – a rapid access to atomic charges. Tetrahedron 36:3219–3228
Sanner MF (1999) Python: a programming language for software integration and development. J Mol Graph Model 17:57–61
Solis FJ, Wets JB (1981) Minimization by random search techniques. Math Oper Res 6:19–30
Huey R, Morris GM, Olson AJ, Goodsell DS (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28:1145–1152
Boehr DD, Wright PE (2008) How do proteins interact? Science 320:1429–1430
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Obiol-Pardo C, Granadino-Roldan JM, Rubio-Martinez J (2008) Protein-protein recognition as a first step towards the inhibition of XIAP and Survivin anti-apoptotic proteins. J Mol Recognit 21:190–204
Pang YP, Xu K, El Yazal J, Prendergast PG (2000) Successful molecular dynamics simulation of the zinc-bound farnesyltransferase using the cationic dummy atom approach. Protein Sci 9:1857–1865
Acknowledgments
We thank Jan Fuhrmann and Dirk Neumann (Center for Bioinformatics, Saarland University) for making available their BALLPass implementation to us.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 4033 kb)
Rights and permissions
About this article
Cite this article
Eyrisch, S., Medina-Franco, J.L. & Helms, V. Transient pockets on XIAP-BIR2: toward the characterization of putative binding sites of small-molecule XIAP inhibitors. J Mol Model 18, 2031–2042 (2012). https://doi.org/10.1007/s00894-011-1217-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-011-1217-y