Abstract
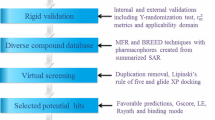
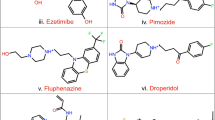
The three-dimensional (3D) structure of the substrate binding domain (SBD) of human ubiquitin ligase Siah2 (seven in absentia homolog) was constructed based on the homology modeling approach using the Modeller 9v7 program. The molecular dynamics method was utilized to refine the model and it was further assessed by ProSA, three-dimensional structural superposition (3d-SS) and PROCHEK in order to analyze the quality and reliability of the generated model. Furthermore, we predicted the binding pocket of Siah2 and also validated it by both blind and normal docking using a known functional inhibitor, menadione. Using structure-based high-throughput virtual screening, we identified five lead drug-like molecules against the modeled SBD of Siah2 and analyzed its pharmacokinetic properties to identify the potential inhibitors for Siah2. The docking results for menadione and the lead molecules at the ligand binding site of SBD of Siah2 revealed that the residue Ser39 (corresponding to Ser167 in the full-length protein) is consistently involved in strong hydrogen bonding, and plays an important role in phosphorylation and the enhanced activity of Siah2.






Similar content being viewed by others
References
Nakayama K (2009) Cellular signal transduction of the hypoxia response. J Biochem 146:757–765
Nakayama K, Qi J, Ronai Z (2009) The ubiquitin ligase Siah2 and the hypoxia response. Mol Cancer Res 7:443–451
House CM, Moller A, Bowtell DD (2009) Siah proteins: novel drug targets in the Ras and hypoxia pathways. Cancer Res 69:8835–8838
Schnell JD, Hicke L (2003) Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem 278:35857–35860
Nakayama K, Frew IJ, Hagensen M, Skals M, Habelhah H, Bhoumik A, Kadoya T, Erdjument-Bromage H, Tempst P, Frappell PB, Bowtell DD, Ronai Z (2004) Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell 117:941–952
House CM, Hancock NC, Moller A, Cromer BA, Fedorov V, Bowtell DD, Parker MW, Polekhina G (2006) Elucidation of the substrate binding site of Siah ubiquitin ligase. Structure 14:695–701
Polekhina G, House CM, Traficante N, Mackay JP, Relaix F, Sassoon DA, Parker MW, Bowtell DD (2002) Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling. Nat Struct Biol 9:68–75
Qi J, Nakayama K, Gaitonde S, Goydos JS, Krajewski S, Eroshkin A, Bar-Sagi D, Bowtell D, Ronai Z (2008) The ubiquitin ligase Siah2 regulates tumorigenesis and metastasis by HIF-dependent and -independent pathways. Proc Natl Acad Sci USA 105:16713–16718
Brizel DM, Scully SP, Harrelson JM, Layfield LJ, Bean JM, Prosnitz LR, Dewhirst MW (1996) Tumor oxygenation predicts for the likelihood of distant metastases in human soft tissue sarcoma. Cancer Res 56:941–943
Moller A, House CM, Wong CS, Scanlon DB, Liu MC, Ronai Z, Bowtell DD (2009) Inhibition of Siah ubiquitin ligase function. Oncogene 28:289–296
Huang J, Tsvetkov L, Qu K, Daniel-Issakani S, Payan DG (2008) Approaches to discovering drugs that regulate E3 ubiquitin ligases. Ernst Schering Found Symp Proc 153–170
Nadeau RJ, Toher JL, Yang X, Kovalenko D, Friesel R (2007) Regulation of Sprouty2 stability by mammalian Seven-in-Absentia homolog 2. J Cell Biochem 100:151–160
Shaw AT, Meissner A, Dowdle JA, Crowley D, Magendantz M, Ouyang C, Parisi T, Rajagopal J, Blank LJ, Bronson RT, Stone JR, Tuveson DA, Jaenisch R, Jacks T (2007) Sprouty-2 regulates oncogenic K-ras in lung development and tumorigenesis. Genes Dev 21:694–707
Seta KA, Spicer Z, Yuan Y, Lu G, Millhorn DE (2002) Responding to hypoxia: lessons from a model cell line. Sci STKE 146:re11
Khurana A, Nakayama K, Williams S, Davis RJ, Mustelin T, Ronai Z (2006) Regulation of the ring finger E3 ligase Siah2 by p38 MAPK. J Biol Chem 281:35316–35326
Emerling BM, Platanias LC, Black E, Nebreda AR, Davis RJ, Chandel NS (2005) Mitochondrial reactive oxygen species activation of p38 mitogen-activated protein kinase is required for hypoxia signaling. Mol Cell Biol 25:4853–4862
Zundel W, Schindler C, Haas-Kogan D, Koong A, Kaper F, Chen E, Gottschalk AR, Ryan HE, Johnson RS, Jefferson AB, Stokoe D, Giaccia AJ (2000) Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev 14:391–396
Shah M, Stebbins JL, Dewing A, Qi J, Pellecchia M, Ronai ZA (2009) Inhibition of Siah2 ubiquitin ligase by vitamin K3 (menadione) attenuates hypoxia and MAPK signaling and blocks melanoma tumorigenesis. Pigment Cell Melanoma Res 22:799–808
McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32:W20–25
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Berman H, Henrick K, Nakamura H (2003) Announcing the worldwide Protein Data Bank. Nat Struct Biol 10:980
Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Aug:2.3
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Sumathi K, Ananthalakshmi P, Roshan MN, Sekar K (2006) 3dSS: 3D structural superposition. Nucleic Acids Res 34:W128–132
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:W407–410
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theor Comput 4:435–447
McDonald NA, Jorgensen WL (1998) Development of an all-atom force field for heterocycles. Properties of liquid pyrrole, furan, diazoles, and oxazoles. J Phys Chem B 102:8049–8059
Jorgensen W, Chandrasekhar J, Madura J, Impey R, Klein M (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Kontoyianni M, McClellan LM, Sokol GS (2003) Evaluation of docking performance: comparative data on docking algorithms. J Med Chem 47:558–565
Schrodinger, LLC (2005) Glide: molecular docking tool, v.4.0. Schrodinger, LLC, New York
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749
Jorgensen WL (2006) QikProp, v.3.0. Schrodinger, LLC, New York
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26
Acknowledgments
We gratefully acknowledge a generous grant from the Lee Foundation, Singapore, for this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Anupriya, G., Roopa, K., Basappa, S. et al. Homology modeling and in silico screening of inhibitors for the substrate binding domain of human Siah2: implications for hypoxia-induced cancers. J Mol Model 17, 3325–3332 (2011). https://doi.org/10.1007/s00894-011-1025-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-011-1025-4