Abstract
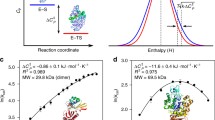
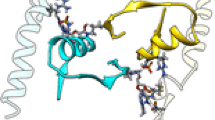
The dependence of some molecular motions in the enzyme 1,3-1,4-β-glucanase from Bacillus licheniformis on temperature changes and the role of the calcium ion in them were explored. For this purpose, two molecular dynamics simulated trajectories along 4 ns at low (300 K) and high (325 K) temperatures were generated by the GROMOS96 package. Several structural and thermodynamic parameters were calculated, including entropy values, solvation energies, and essential dynamics (ED). In addition, thermoinactivation experiments to study the influence of the calcium ion and some residues on the activity were conducted. The results showed the release of the calcium ion, which, in turn, significantly affected the movements of loops 1, 2, and 3, as shown by essential dynamics. These movements differ at low and high temperatures and affect dramatically the activity of the enzyme, as observed by thermoinactivation studies.







Similar content being viewed by others
Abbreviations
- MD:
-
Molecular dynamics
- RF:
-
Reaction field
- RMSD:
-
Root mean square deviation
- ED:
-
Essential dynamics
References
Planas A (2000) Biochim Biophys Acta 1543:361–382
Juncosa M, Pons J, Dot T, Querol E, Planas A (1994) J Biol Chem 269:14530–14535
Malet C, Vallés J, Bou J, Planas A (1996) J Biotechnol 48:209–219
Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon J, Davies G (1995) Proc Natl Acad Sci USA 92:7090–7094
Planas A, Juncosa M, Lloberas J, Querol E (1992) FEBS Lett 308:141–145
Welfle K, Misselwitz R, Welfle H, Politz O, Borriss R (1994) J Biomol Struct Dyn 11:1417–1424
Welfle K, Misselwitz R, Welfle H, Politz O, Borriss R (1995) Eur J Biochem 229:726–735
Welfle K, Misselwitz R, Politz O, Borriss R, Welfle H (1996) Protein Sci 5:2255–2265
Keitel T, Meldgaard M, Heinemann U (1994) Eur J Biochem 222:203–214
Lloberas J, Querol E, Bernués J (1988) Appl Microbiol Biotechnol 29:32–38
Gargallo R, Hünenberger P, Aviles F, Oliva B (2003) Protein Sci 10:2161–2172
Amadei A, Linssen A, Berendsen H (1993) Proteins Struct Funct Genet 17:412–425
Yang C, Gouri S, Kuczera K (2001) J Biomol Struct Dyn 19:247–271
van Gunsteren W, Billeter S, Eising A, Hünenberger P, Früger P, Mark A, Scott W, Tironi I (1996) Biomolecular simulation: the GROMOS96 manual and user guide. Verlag der Fachvereine, Zürich
Berendsen H, Grigera J, Straatsma T (1987) J Phys Chem 91:6269–6271
Rickaert J, Ciccotti G, Berendsen H (1977) J Comput Chem 23:327–341
Tironi I, Sperb R, Smith P, van Gunsteren W (1995) J Chem Phys 102:5451–5459
Hünenberger, P, van Gunsteren W (1998) J Chem Phys 108:6117–6134
Gargallo R, Oliva B, Querol E, Avilés F (2000) Protein Eng 13:21–26
Richmond T (1984) J Mol Biol 176:63–89
Nicholls A, Honing B (1991) J Comput Chem 12:435–440
Bashford D (1997) An object-oriented programming suite for electrostatic effects in biological molecules. In: Ishikawa Y, Reynders J, Tholburn M (eds) Scientific computing in object-oriented parallel environments. Springer, Berlin Heidelberg New York, pp 233–240
Schlitter J (1993) Chem Phys Lett 215:617–621
Schäfer H, Mark A, van Gunsteren W (2000) J Chem Phys 113:7809–7817
Schäfer H, Daura X, Mark AE, van Gunsteren W (2001) Proteins 43:45–56
Schäfer H, Smith LJ, Mark A, van Gunsteren W (2002) Proteins 46:215–224
Sherer E, Harris S, Soliva R, Orozco M, Laughton C (1999) J Am Chem Soc 121:5981–5991
Pons J, Planas A, Juncosa M, Querol E (1997) Methods Mol Biol 67:209–218
Malet C, Viladot J, Ochoa A, Gállego B, Brosa C, Planas A (1995) Carbohydr Res 274:285–301
Mozo-Villarias A, Cedano J, Querol E (2003) Protein Eng 16:279–286
Acknowledgements
RG received a grant from the Spanish MEC (BQU2003-00191). BO received grants from the Fundación Ramón Areces and from the Spanish MEC (BIO2002-03609 and BIO2005-00533). EQ received grants from Ministerio de Ciencia y Tecnologia (BIO2001-264) and from the Centre de Referència de R+D de Biotecnología de la Generalitat de Catalunya.
Author information
Authors and Affiliations
Corresponding author
Additional information
The first two authors contributed equally to this work
Rights and permissions
About this article
Cite this article
Gargallo, R., Cedano, J., Mozo-Villarias, A. et al. Study of the influence of temperature on the dynamics of the catalytic cleft in 1,3-1,4-β-glucanase by molecular dynamics simulations. J Mol Model 12, 835–845 (2006). https://doi.org/10.1007/s00894-006-0110-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-006-0110-6