Abstract
Objective
To identify the characteristics of the oral microbiota and the relationship of the dental caries and periodontal status in patients aged 0 to 18 years with non-syndromic cleft lip and palate (CLP).
Materials and methods
A systematic review of the literature was carried out. Five databases were consulted, including publications in English, Spanish and Portuguese. The evaluations of the quality of the observational studies and the experimental studies were carried out with the Newcastle–Ottawa scale and CONSORT guidelines, respectively. The risk of bias of the studies was determined using Rev Manager 5.4, and 5 publications were meta-analyzed.
Results
The cariogenic microbiota of children and adolescents with cleft lip and palate was similar to that of children without clefts, although with higher counts of Streptococcus mutans and Lactobacillus spp. The periodontopathogenic microbiota was related to the presence of Campylobacter spp, Fusobacterium spp, Fusobacterium nucleatum, Prevotella intermedia/nigrescens, Parvimonas micra and Porphyromonas gingivalis, considered microorganisms with high pathogenic capacity. Heterogeneity was shown in relation to the microbiota and the type of fissure, presenting numerous microorganisms associated with the pre- and post-surgical condition (cheilorrhaphy and palatorrhaphy) such as Staphylococcus aureus, Streptococcus beta hemolyticus, Klebsiella pneumoniae and Klebsiella oxytoca, Moraxella catarrhalis, Candida spp, Candida albicans, Candida krusei and Candida tropicalis. The meta-analysis revealed that patients with cleft lip and palate were 2.03 times more likely to have caries than the control group (p<0.005).
Conclusion
In the microbiota, there was a great diversity of microorganisms that can vary according to the type of fissure and surgical interventions predisposing patients to a greater probability of dental caries, it is important to take into account the technique used to describe the oral microbiota in order to be able to compare the different studies.
Clinical relevance
Studying the microbiota and the relationship of dental caries and periodontal status in children and adolescents with cleft lip and palate can facilitate the comprehensive care of patients with these conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cleft lip and palate (CLP) is one of the most frequent congenital craniofacial malformations and originates between the fourth and sixth week of intrauterine life [1, 2]. The World Health Organization (WHO) reports a worldwide yearly incidence of CLP of 1 in every 700 live births [3]. In South America, the prevalence has been reported to be 1 in 800 live births, and in Colombia, according to the fourth national study of oral health in Colombia (ENSAB IV 2013-2014), the prevalence is 1 in 700 (0.07%) [3].
This malformation develops as an anatomical defect in the fissured area, contributing to problems with phonation, chewing, and swallowing, malposition and dental alterations in shape, number and structure; this discontinuity of the tissues of the oral and nasal cavities contributes to the formation of unbalanced bacterial ecosystems [1, 4,5,6]. In the first months of life, surgical interventions are generally performed to repair the defect through primary closure of the fissure, for which the literature has shown [3] a higher prevalence of gram-negative microorganisms before surgery and a higher frequency of gram-positive microorganisms after surgery, related to the surgical closure of the nasopharyngeal space. Furthermore, the risk of early colonization of microorganisms increases significantly in children with CLP [3].
The microbiota in patients with CLP has been extensively investigated, and it has been suggested that children and adolescents with CLP may have elevated levels of Streptococcus mutans, Candida spp. and Lactobacillus spp. in saliva before and after cheilorrhaphy and palatorrhaphy [3, 7]. In some CLP patients, beta hemolytic Streptococcus and Staphylococcus aureus have been isolated from the nasal cavity [2], and the presence of Candida spp. has been shown to be related to the immunosuppression present at the time of birth [8].
Nonsyndromic cleft lip and palate (NSCLP) predisposes patients to the formation of retentive areas of biofilm, and as sequelae of surgical intervention, recurrent oronasal fistulas, wound dehiscences and scarring flanges appear, generating retention zones, which together cause different pH values, local oxygen concentrations, redox states, ionic compositions, buffering capacity and mechanical interactions, which, after the accumulation of food and oral and nasal fluids, create an environment conducive to the growth of various bacterial groups [1, 7]. Changes in the microbiota can produce alterations in the ecological balance, causing environmental disturbances that lead to a predominance of harmful microorganisms that contribute to the pathogenesis of oral diseases such as tooth decay and periodontal disease, among other pathologies [1, 9].
In this sense, different authors mention that the oral microbiota is different in children who present this condition than in children who do not present it [10, 11]. However, a consensus has not yet been established regarding the characterization of the oral microbiota in this population and whether it differs from the microbiota of the population without NSCLP. Therefore, the aim of this study was to identify the characteristics of the oral microbiota and the relationship of the oral microbiota with dental and periodontal status in patients aged 0 to 18 years with NSCLP.
Materials and methods
A systematic review of the literature was performed in accordance with the PRISMA 12 guidelines [12]. The following question was posed under the PICO structure: P Children and adolescents aged 0 to 18 years with non-syndromic cleft lip and palate; I Oral microbiota; C Children and adolescents aged 0 to 18 years without non-syndromic cleft lip and palate; O Dental and periodontal status.
What are the characteristics of the oral microbiota in children and adolescents aged 0 to 18 years with non-syndromic cleft lip and palate and the relationship of the oral microbiota with dental and periodontal status?
Eligibility criteria
Studies that evaluated the oral microbiota through analytical observational designs and clinical trials were included. Articles that included patients had received antimicrobial therapy were excluded.
Search strategy
Electronic searches were carried out in Medline-PubMed, Embase-Elsevier, EBSCO, Scopus, and Web of Science, in addition to a gray literature search in Google Scholar. The search was restricted to articles in English, Spanish and Portuguese published from January 1, 1985, to June 30, 2020. Search descriptors were used in controlled and uncontrolled language related to cleft lip, cleft palate, cleft lip and palate no syndromic, microbiology, biofilm, dental caries, periodontal diseases, dental caries susceptibility and uncontrolled language such as child, children, teenagers, oral microbiota, oral microbiome, periodontal state, and dental state. Different combinations of terms were used in the search strategies through the Boolean operators AND, OR and NOT; the search strategies were tailored to the particularities of each database.
Methodological evaluation of the publications
Assessment of the methodological quality of the publications was carried out through the Newcastle–Ottawa scale (NOS) [13] for analytical observational studies and the Consort checklist for clinical trial.
Statistical and analytical aspects
The search and extraction of the information was carried out independently by 4 reviewers. Once the data were extracted from the articles, they were analyzed using Review Manager 5.4 software.
To calculate the effect size, the articles that included ORs with the 95% CIs were taken into account, as well as the raw data for the studies with an analytical observational design. Means were analyzed to determine differences; heterogeneity was assessed using the Q statistical method based on v2 and I2, with significance indicated by P < 0.05.
Results
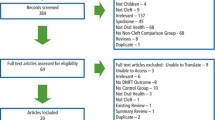
The flowchart for article inclusion is shown in Fig. 1.
The essential information for each study is summarized in Table 1
Risk of bias analysis of the studies included in the meta-analysis
Using Rev Manager 5.4, the risk of bias in the included studies was evaluated; the results indicated that the majority of the studies (90%) had a prospective longitudinal case–control design in which there was a high level of bias (Fig. 2).
Cariogenic microbiota and its relationship with dental status
Regarding the cariogenic microbiota in lip-palate sites, there are microorganisms similar to those found in patients without clefts, although with differences in the percentage. In studies by Ahluwalia et al. [14], Lucas et al. [15], and Hassani et al. [22], there were significantly higher counts of Streptococcus mutans and Lactobacillus spp. in patients with CLP. Cheng et al. [20] and Sundell et al. [29] reported significantly higher percentages of Lactobacillus spp. in patients with this condition. In addition, microorganisms related to endodontic lesions, such as Peptostreptococcus micros and Catonella morbi, were detected.
Regarding dental status, Ahluwalia et al. [14] reported higher CPOD-ceod index scores (decayed, lost or filled teeth) in CLP patients than in patients in the control group, and Lucas et al. [15] and Durhan et al. [16] did not find statistically significant DMFT index scores for patients with CLP.
Periodontopathogenic microbiota and its relationship with the periodontal state.
A characterization of the pathogenic microbiota of the periodontium in patients with CLP is presented as reported by Rawashdeh et al.8 Costa et al.10 Perdikogianni et al. [11], Ahluwalia et al. [14], Funahashi et al. [21], and Quirynen et al. [25]: Capnocytophaga gingivalis, Eikenella Corrodens, Wolinella spp. Actinomyces spp , Campylobacter spp, Fusobacterium spp, Fusobacterium nucleatum, Prevotella intermedia/nigrescens, Peptostreptococcus micros and Porphyromonas gingivalis.
Regarding the gingival and periodontal condition, Ahluwalia et al. [14], Rawashdeh et al. [8], Funahashi et al. [21], Perdikogianni et al. [11], Quirynen et al. [25], and Costa et al. [10] reported a higher gingival index, biofilm index, probing depth, bleeding on probing and loss of insertion in patients with CLP, and Lucas et al. [15] did not find significant differences in periodontal indicators between patients with and without CLP.
Oral microbiota and its relationship with type of fissure
There is heterogeneity in relation to the microbiota and type of fissure. Rawashdeh et al. [8], who analyzed of samples of tongue, nasal and palatal mucosa, reported a colonization rate by Candida spp. that was higher in patients with bilateral cleft lip and palate (77.7%) than in patients with unilateral cleft lip and palate (57, 1%), a finding that was attributed to the fact that patients with clefts have poor oral hygiene.
Bokhout et al. [24], who analyzed saliva and dental plaque microbiological cultures, reported that patients with bilateral cleft lip and palate had a higher percentage of microorganisms: Streptococcus mutans (in saliva by 57.1% and in teeth by 71.4%) and Lactobacillus spp. (in saliva and teeth by 14.3%); however, they reported a lower percentage of these microorganisms in isolated labial fissures and isolated cleft palate.
Streptococcus mitis, Streptococcus salivarius, Staphylococcus aureus MSSA, Staphylococcus epidermidis, Enterobacter cloacae, Klebsiella pneumoniae and Klebsiella oxytoca were more predominant in CLP than cleft soft palate.
Zhang et al.2who analyzed saliva and nasal samples through microbial genomic DNA and PCR, revealed that the genera Lautropia spp. and Bacillus spp. were less abundant in saliva samples from individuals with a complete cleft palate (p = .029).
Oral microbiota and its relationship with surgical intervention
Studies have identified microorganisms associated with the presurgical and postsurgical conditions related to cheilorrhaphy and palatorrhaphy. Cocco et al. [17], Tuna et al. [18], Arief et al. [19], Thomas et al. [27], Hupkens et al. [28], and Machorowska et al. [1] identified different percentages of Staphylococcus aureus in patients with CLP. Tuna et al. [18] reported that the transmission of this microorganism increases with the size of the postsurgical residual oronasal fistula. Cocco et al. [17], Hupkens et al. [28], and Thomas et al. [27] reported that beta-hemolytic Streptococcus (Streptococcus pyogenes) was associated with a high risk of complications such as dehiscence of the surgical wound. Cocco et al. [17], Machorowska et al. [1], and Hupkens et al. [28] isolated the genera Klebsiella pneumoniae and Klebsiella oxytoca, which were more predominant in the preoperative period and decreased in proportion after surgery.
Thomas et al. [27] and Hupkens et al. [28] isolated Moraxella catarrhalis in patients who underwent surgery, and Machorowska et al. [1] and Cocco et al. [17] isolated methicillin-resistant Staphylococcus aureus, which increased in number after surgical repair. Rawashdeh et al. [8] and Da Silva et al. [26] found Candida spp., Candida albicans, Candida krusei and Candida tropicalis before and after surgical repair, reporting higher proportions of this microorganism after surgical intervention, with statistically significant differences, noting that the greater was the number of surgical interventions, the greater the colonization by Candida spp.
Liu et al. [9] found significant variations in the microbiota in patients undergoing surgery who presented inflammation compared to those without inflammation, including the following: operational taxonomic units (OTUs) related to inflammation – Tannerella spp., Porphyromonas spp., Gemella spp., Moraxella spp., Prevotella nigrescens and Prevotella intermedia; and related OTUs without the presence of inflammation – Lautropia spp., Neisseria spp., Capnocytophaga spp., Veillonella dispar, Veillonella parvula and Prevotella melaninogenica. OTUs corresponding to Streptococcus spp. and Prevotella spp. were present in both groups.
Quantitative analysis
Results of the meta-analysis
A meta-analysis was performed to determine the association between the microorganisms analyzed and dental caries. Of the 23 articles included in this review, 5 were meta-analyzed.
Streptococcus mutans
Two studies were analyzed that reported the outcome in means of the presence of Streptococcus mutans, and that there was no difference between the means for healthy patients and those with CLP. The results of these studies show very high heterogeneity, suggesting this analysis should be carefully reviewed (Fig. 3).
Lactobacillus
For the outcome of the presence of Lactobacillus spp., there were no significant differences between patients with CLP and healthy patients, and the diversity of the studies led to high heterogeneity; however, the p value was significant (Fig. 4).
Caries risk
For the analysis of caries risk in patients with CLP and healthy patients, 3 studies were included that evaluated the proportion of patients with caries in both groups. With the 3 included studies, the OR was 2.03, indicating that patients with CLP were 2.03 times more likely to have cavities than were patients in the control group. Heterogeneity was low (p < 0.005) (Fig. 5).
Discussion
Children with CLP have a very diverse microbiome and it has now been reported that polymicrobial communities induce a dysregulated and destructive host response through a global mechanism termed polymicrobial synergy and dysbiosis. Microorganisms in the communities tend to interact synergistically to enhance colonization, persistence or pathogenicity [30].
Regarding the cariogenic microbiota in CLP sites, the results of this investigation showed that there are microorganisms similar to those found in healthy patients, although with a higher percentage in fissure sites, especially higher counts of Streptococcus mutans and Lactobacillus spp. These findings are like those reported by Chaudhari et al. [31], who evaluated the Streptococcus and Lactobacillus spp. Counts in saliva and reported higher counts in patients with cleft as well as increased salivary Lactobacillus spp. counts that were higher (60%) in children with CLP. Similar results were reported by Parapanisiou et al. [32], who reported elevated levels of Lactobacillus spp. (> 10 5 CFU/ml) in patients with CLP.
Contrary to the findings reported in the present investigation, Shashni et al.7found no statistically significant difference in terms of Lactobacillus spp. levels among children with CLP, children with a high risk of caries without cleft and children without cavities and without cleft. Likewise, Dahllöf et al. [33] did not observe differences in the salivary count of Lactobacillus spp. in groups with cleft lip and cleft palate.
The higher percentage of Streptococcus spp. and Lactobacillus spp. could be related to the anatomical and treatment conditions that patients with CLP present. Shelton et al. [34] reported that presurgical orthopedic devices alter the conditions of the oral cavity, establishing acidic environments as a result. The acrylic material of the plate generally presents roughness, which increases the probability of colonization of Lactobacillus spp. and consequently increases Streptococcus spp. [34].
In relation to the dental condition, DMFT-ceod index scores were higher for these patients than for the control group. Likewise, patients with CLP had a 2.03× greater probability of presenting caries than did the control group (p < 0.005). These findings do not reveal a direct relationship with dental caries; however, the microbiota is an essential biological factor for the development of lesions, its composition is dynamic, and its evolution depends greatly on the intake of sugars and the use of fluoride [18, 20, 22]
These results are similar to those reported by Chaudhari et al. [31], who analyzed the presence of dental caries in patients with and without CLP, reporting an increase in the number of teeth with caries (DMFT scores that increased from 2-3 to 4-6), which was significantly correlated with counts of salivary Lactobacillus spp. both in children with CLP and in non-cleft children; there was no significant correlation with counts of salivary Streptococcus spp. in children with and without CLP.
Worth et al. [35], in a systematic review and meta-analysis, reported that the overall pooled mean difference in the ceod was 0.63 (95% CI: 0.47 to 0.79) and in the DMFT was 0.28 (95% CI: 0.22 to 0.34), suggesting that individuals with cleft lip and/or palate have a higher frequency of dental caries in both primary and permanent teeth. Contrary to our results, Bastos et al. [36], in a Brazilian population, found no significant differences in dental condition between children with and without CLP.
Several factors could influence the risk of caries in patients with CLP. Allam et al. [37] analyzed primary and mixed dentition in patients with CLP and found a direct correlation between caries, the intake of foods containing sugar between meals and hygiene habits. They also reported that there was a direct correlation between CPOD-ceod index scores and a higher intake of sugary foods.
Regarding the periodontal microbiota, Campylobacter spp., Fusobacterium spp., Fusobacterium nucleatum, Prevotella intermedia/nigrescens, Parvimonas micra and Porphyromonas gingivalis have been isolated in patients with CLP and are considered microorganisms of great pathogenic capacity [8, 10, 11, 14, 15, 25] The presence of periodontopathogenic bacteria such as Porphyromonas gingivalis is relevant, an etiological agent in severe forms of periodontitis, which is not a common disease in patients under 18 years of age; its presence in children and adolescents has been related to immunological alterations as a modification in neutrophil chemotaxis. Porphyromonas gingivalis can locally invade periodontal tissues and evade host defense mechanisms. In doing so, it uses virulence factors that cause the dysregulation of innate immune and inflammatory responses [38].
Lamont et al [39] reported that in periodontal diseases, polymicrobial communities induce a dysregulated and destructive host response through a global mechanism termed polymicrobial synergy and dysbiosis. In contrary to what occurs in the gastrointestinal tract, periodontal diseases are associated with an increase in microbiome diversity, which is thought to be a consequence of additional nutrients derived from host tissue damage and increased physical space as the gingival cleft deepens.
Mombelli et al. [40] analyzed the microbiota in patients with unilateral and bilateral CLP and observed the presence of gram-negative anaerobic microorganisms. They also reported the presence of Fusobacterium spp., Prevotella melaninogenica and Prevotella intermedia in patients with CLP, but they did not detect Porphyromonas gingivalis, Actinobacillus spp. and Aggregatibacter actinomycetemcomitans in the study population.
Weckwerth et al. [41] conducted a study with 31 patients with CLP and chronic suppurative otitis media and obtained positive cultures from 83% of the patients. Pseudomonas aeruginosa (54.9%), Staphylococcus aureus (25.9%) and Enterococcus faecalis (19.2%) were isolated, but no anaerobes were isolated by culture, and the polymerase chain reaction assays revealed 1 or more bacteria in 97.1% of the samples. Anaerobic microorganisms were detected by polymerase chain reaction assays, for example, Fusobacterium nucleatum, Bacteroides fragilis and Peptostreptococcus anaerobius. This finding suggests that patients with this condition present communication between the ear and the oral cavity, and through this route, there could be an exchange of microorganisms.
In this review, 6 studies identified that patients with CLP presented higher gingival index values and biofilm index values, deeper probing depth, and more bleeding on probing and attachment loss. These results support the results reported by Parapanisiou et al. [32], who found that the biofilm index was significantly higher in patients with CLP than in the control group (p = 0.0003). Likewise, Veiga et al [42], in a study with 156 children between 5 and 18 years of age with CLP, showed that fissured patients presented a higher plaque index and gingival index and greater depth when probing.
Plakwicz et al. [43] evaluated the periodontal index in 34 patients with a divided mouth, reporting that the depth of probing and the loss of clinical attachment were greater in the lateral incisors and canines adjacent to the cleft lip than in the same contralateral teeth without the fissure. Wyrębek et al. [44] analyzed 15 patients aged 6 to 18 years with bilateral clefts and found greater bleeding on probing and loss of attachment in the teeth adjacent to the cleft.
The publications analyzed in this study identified microorganisms associated with the pre- and postsurgical conditions related to cheilorrhaphy and palatorrhaphy. Studies had reported a significantly higher count of Staphylococcus aureus in saliva samples from children with larger oronasal fistulae and indicated a positive correlation between the size of the fistula and the frequency of transmission of Staphylococcus aureus to an oral environment. Adeyemo et al. [45] reported that Staphylococcus spp. is a commensal of the skin and nose and that cleft surgery involves both extraoral and intraoral incisions that often lead to communication between the skin and nasal mucosa. Therefore, contamination of the surgical wound in patients with CLP and the subsequent entry of this microorganism into the bloodstream may explain the high prevalence of Staphylococcus spp. observed in this study. Authors such as Chuo and Timmons [46] conclude that children with unrepaired CLP have an increased risk of carrying Staphylococcus aureus and that these risks should be taken into account when choosing the relevant preoperative and postoperative bacteriology tests.
The oral microbiota is highly diverse, consisting of hundreds of bacterial species in the different oral microenvironments, and they play an important role in determining the state of health or disease in the host [47,48,49]. However, due to their high complexity and the limitations of the methodological tools available to describe the microorganisms associated with health or disease, together with the fact that more than one third of oral bacteria are not culturable, traditional microbiological approaches provide incomplete information on the natural communities that inhabit the oral cavity [50, 51].
In this systematic literature review and meta-analysis, it was observed that of the 23 articles included, 14 (61%) performed microbiological cultures to search for bacteria and 2 (8.7%) of the studies for the identification of fungi, specifically Candida species, 3 (13%) used commercial techniques with pre-established panels of microorganisms such as the CRT test system and Dentocult®, Likewise, 2 (8.7%) performed molecular biology assays, one of them DNA-DNA hybridization and the other PCR-DGGE in which a limited number of bacterial species are analyzed and only 2 (8.7%) of the studies included new generation sequencing techniques or NGS, by amplification of a fragment of 16S rRNA.
PCR amplification of fragments of the gene encoding the 16S rRNA and subsequent sequencing permits the identification of bacterial genera and in some cases even allows the determination of species, depending on the bacterial genus evaluated and the variable region analyzed. This gene has an approximate size of 1500 nucleotides and has 9 variable regions. In studies of microbiota associated to human infectious diseases, the most used region for its analysis is V3-V4 [52].
Recently, sequencing of the entire microbiome including bacteria, archaea, fungi, parasites and DNA-type viruses is being used in different pathologies of medical importance by sequencing all the DNA present in the sample; however, the bioinformatic analysis of all this information obtained is extremely laborious, but it allows us to describe species, subspecies or even strains present, in addition to providing information on the abundance in which they are present. However, to date, it has not been possible to find metagenomic studies that describe the different microbiological profiles in patients with and without NSCLP. Researchers are even proposing to integrate information derived from the metagenome and metatranscriptome simultaneously to truly understand the role of the microbiota in disease generation. The metagenome describes qualitatively and quantitatively the totality of bacteria present in different niches, including those of the oral cavity. The metatranscriptome, on the other hand, provides a profile of bacterial transcripts also qualitatively and quantitatively reflecting the transcriptional activity of all the microorganisms present in the niches under study. These transcripts could reflect the presence of bacterial metabolites.
It is important to mention that the type of sample makes it difficult to compare the studies, since the bacterial community of the subgingival and supragingival microbiota, although they tend to be similar, is different from that found in the mucosa and that found in saliva; in fact, some oral bacteria show a specific tropism towards the different biological surfaces in the oral cavity [53]. For example, Streptococcus salivarius, S. oralis, S. constellatus, S. mitis, S. intermedius and S. anginosus are preferentially found in soft tissues and saliva, compared to Streptococcus sanguis which preferentially colonizes dental surfaces in particular supragingival plaque.
Conclusions
-
The present investigation identified that patients with CLP had higher counts of Streptococcus mutans and Lactobacillus spp.
-
The results of the meta-analysis suggest that individuals with CLP may have greater risk of developing dental caries; therefore, this risk should be taken into account when making clinical decisions and adopting preventive measures to reduce oral comorbidities in these patients.
-
Periodontopathogenic bacteria were observed in fissure areas, highlighting the presence of Porphyromonas gingivalis.
-
The sites adjacent to a fissure have higher plaque index and gingival index values, a deeper probing depth and greater loss of attachment.
-
The larger the oronasal fistula is, the greater the percentage of Staphylococcus spp.
Recommendations
The description of the oral microbiota should be carefully interpreted as it is a function of the methodology used, mainly because traditional microbiological cultures present limitations as they only describe that microbiota that can be cultured. We recommend that future studies incorporate a single sample collection method or unify the type of sample. In addition, large-scale clinical studies should be conducted. Metagenomics and metatranscriptomics studies are recommended in children and adolescents with and without non-syndromic cleft lip and palate.
Limitations
Future studies should incorporate the units of measure for microorganisms and adequately describe their different methods of sample collection to unify knowledge of this topic. Additionally, large-scale clinical studies should be conducted.
Data Availability
The datasets and studies used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CLP:
-
Cleft lip and palate
- NSCLP:
-
Nonsyndromic cleft lip and palate
- WHO:
-
World Health Organization
- ENSAB:
-
National study of oral health
- SSP:
-
Species
References
MacHorowska-Pieniazek A, Mertas A, Skucha-Nowak M, Tanasiewicz M, Morawiec T (2017) A comparative study of oral microbiota in infants with complete cleft lip and palate or cleft soft palate. BioMed Res Int. https://doi.org/10.1155/2017/1460243
Zhang M, Wang R, Liao Y, Buijs MJ, Li J (2016) Profiling of oral and nasal microbiome in children with cleft palate. Cleft Palate-Craniofacial J 53(3):332–338. https://doi.org/10.1597/14-162
Durhan MA, Topcuoglu N, Kulekci G, Ozgentas E, Tanboga I (2019) Microbial Profile and Dental Caries in Cleft Lip and Palate Babies Between 0 and 3 Years Old. Cleft Palate Craniofac J 56(3):349–356. https://doi.org/10.1177/1055665618776428
Rengifo Reina HA (2016) Dental characterization of colombian children with non syndromic cleft lip and palate. Rev Odontológica Mex 20(3):e175–e181. https://doi.org/10.1016/j.rodmex.2016.08.014
Gómez O, Puerto B (2017) Cleft Lip and Palate. Obstet Imaging Fetal Diagnosis Care Second Ed 45(4):311–316.e1. https://doi.org/10.1016/B978-0-323-44548-1.00065-6
Sundell AL, Ullbro C, Dahlén G, Marcusson A, Twetman S (2018) Salivary microbial profiles in 5-year old children with oral clefts: a comparative study. Eur Arch Paediatr Dent 19(1):57–60. https://doi.org/10.1007/s40368-018-0326-z
Shashni R, Goyal A, Gauba K, Utreja AK, Ray P, Jena AK (2015) Comparison of risk indicators of dental caries in children with and without cleft lip and palate deformities. Contemp Clin Dent 6(1):58–62. https://doi.org/10.4103/0976-237X.149293
Rawashdeh MA, Ayesh JAM, Darwazeh AMG (2011) Oral candidal colonization in cleft patients as a function of age, gender, surgery, type of cleft, and oral health. J Oral Maxillofac Surg 69(4):1207–1213. https://doi.org/10.1016/j.joms.2010.02.044
Liu L, Zhang Q, Lin J et al (2016) Investigating oral microbiome profiles in children with cleft lip and palate for prognosis of alveolar bone grafting. PLoS One 11(5):1–13. https://doi.org/10.1371/journal.pone.0155683
Costa B, De Oliveira Lima JE, Gomide MR, Da Silva Rosa OP (2003) Clinical and Microbiological Evaluation of the Periodontal Status of Children with Unilateral Complete Cleft Lip and Palate. Cleft Palate-Craniofacial J 40(6):585–589. https://doi.org/10.1597/01-083
Perdikogianni H, Papaioannou W, Nakou M, Oulis C, Papagiannoulis L (2009) Periodontal and microbiological parameters in children and adolescents with cleft lip and/or palate. Int J Paediatr Dent 19(6):455–467. https://doi.org/10.1111/j.1365-263X.2009.01020.x
Liberati A, Altman DG, Tetzlaff J et al (2009) The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Health Care Interventions. Expl Elaboration 62. https://doi.org/10.1016/j.jclinepi.2009.06.006
Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25(9):603–605. https://doi.org/10.1007/s10654-010-9491-z
Ahluwalia M, Brailsford SR, Tarelli E et al (2004) Dental caries, oral hygiene, and oral clearance in children with craniofacial disorders. J Dent Res 83(2):175–179. https://doi.org/10.1177/154405910408300218
Lucas VS, Gupta R, Ololade O, Gelbier M, Roberts GJ (2000) Dental health indices and caries associated microflora in children with unilateral cleft lip and palate. Cleft Palate-Craniofacial J 37(5):447–452. https://doi.org/10.1597/1545-1569(2000)037<0447:DHIACA>2.0.CO;2
Durhan MA, Kulekci G, Tanboga I, Ozgentas E, Topcuoglu N (2018) Microbial Profile and Dental Caries in Cleft Lip and Palate Babies Between 0 and 3 Years Old. Cleft Palate-Craniofacial J:105566561877642. https://doi.org/10.1177/1055665618776428
Cocco JF, Antonetti JW, Burns JL, Heggers JP, Blackwell SJ (2010) Characterization of the nasal, sublingual, and oropharyngeal mucosa microbiota in cleft lip and palate individuals before and after surgical repair. Cleft Palate-Craniofacial J 47(2):151–155. https://doi.org/10.1597/08-187.1
Tuna EB, Topçuoglu N, Ilhan B, Gençay K, Kulekçi G (2008) Staphylococcus aureus transmission through oronasal fistula in children with cleft lip and palate. Cleft Palate-Craniofacial J 45(5):477–480. https://doi.org/10.1597/06-247.1
Arief EM, Mohamed Z, Idris FM (2005) Study of viridans streptocci and Staphyloccus species in cleft lip and palate patients before and after surgery. Cleft Palate-Craniofacial J 42(3):277–279. https://doi.org/10.1597/04-083R.1
Cheng LL, Moor SL, Kravchuk O, Meyers IA, Ho CTC (2007) Bacteria and salivary profile of adolescents with and without cleft lip and/or palate undergoing orthodontic treatment. Aust Dent J 52(4):315–321. https://doi.org/10.1111/j.1834-7819.2007.tb00508.x
Funahashi K, Shiba T, Watanabe T et al (2019) Functional dysbiosis within dental plaque microbiota in cleft lip and palate patients. Prog Orthod 20(1):1–11. https://doi.org/10.1186/s40510-019-0265-1
Hassani H, Chen JW, Zhang W, Hamra W (2020) Comparison of Microbial Activity Among Infants With or Without Using Presurgical Nasoalveolar Molding Appliance. Cleft Palate-Craniofacial J 57(6):762–769. https://doi.org/10.1177/1055665620908150
Loveren C, Buijis J, Prahl-andersen B, Ten Catae J (1998) Incidence of mutans streptococci and lactobacilli in oral cleft children wearing acrylic plates from shortly after birth. Oral Microbiol Immunol 13(5):296–291
Bokhout B, Van Loveren C, Hofman FXWM, Buijs JF, Van Limbeek J, Prahl-Andersen B (1996) Prevalence of Streptococcus mutans and lactobacilli in 18-month-old children with cleft lip and/or palate. Cleft Palate-Craniofacial J 33(5):424–428. https://doi.org/10.1597/1545-1569(1996)033<0424:POSMAL>2.3.CO;2
Quirynen M, Dewinter G, Avontroodt P, Heidbüchel K, Verdonck ACC (2003) A split-mouth study on periodontal and microbial parameters in children with complete unilateral cleft lip and palate. J Clin Periodontol 30(1):49–56. https://doi.org/10.1034/j.1600-051X.2003.300108.x
da Silva JJ, da Silva TA, de Almeida H et al (2018) Candida species biotypes in the oral cavity of infants and children with orofacial clefts under surgical rehabilitation. Microb Pathog 124(July):203–215. https://doi.org/10.1016/j.micpath.2018.08.042
Thomas GPL, Sibley J, Goodacre TEE, Cadier MM (2012) The value of microbiological screening in cleft lip and palate surgery. Cleft Palate-Craniofacial J 49(6):708–713. https://doi.org/10.1597/11-063
Hupkens P, Lauret GJ, Dubelaar IJM, Hartman EHM, Spauwen PHM (2007) Prevention of wound dehiscence in palatal surgery by preoperative identification of group A Streptococcus and Staphylococcus aureus. Eur J Plast Surg 29(7):321–325. https://doi.org/10.1007/s00238-007-0116-z
Sundell AL, Ullbro C, Marcusson A, Twetman S (2015) Comparing caries risk profiles between 5- and 10- year-old children with cleft lip and/or palate and non-cleft controls. BMC Oral Health 15(1):1–6. https://doi.org/10.1186/s12903-015-0067-x
Hajishengallis G, Lamont RJ (2012) Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol 27(6):409–419. https://doi.org/10.1111/j.2041-1014.2012.00663.x
Chaudhari PK, Kharbanda OP, Chaudhry R et al (2020) Factors Affecting High Caries Risk in Children With and Without Cleft Lip and/or Palate: A Cross-Sectional Study. Cleft Palate-Craniofacial J:(406). https://doi.org/10.1177/1055665620980206
Parapanisiou V, Gizani S, Makou M, Papagiannoulis L (2009) Oral health status and behaviour of Greek patients with cleft lip and palate. J Eur Acad Paediatr Dent 10(2):85–89. https://doi.org/10.1007/BF03321606
Dahllöf G, Joandi R, Ideberg M, Modeer T (1989) Caries, gingivitis and dental abnormalities in preschool children with cleft lip and/or palate. Fogorv Sz 26(3):233–237
Shelton K (2017) Bacterial Flora in Cleft Lip and Palate Patients Undergoing Presurgical Nasoalveolar Molding Appliance Therapy. ProQuest Diss Theses, p 100. https://search.proquest.com/docview/1937904104?accountid=26642%0Ahttp://link.periodicos.capes.gov.br/sfxlcl41?url_ver=Z39.88-2004&rft_val_fmt=info:ofi/fmt:kev:mtx:dissertation&genre=dissertations+%26+theses&sid=ProQ:ProQuest+Dissertations+%26+Theses+Globa
Worth V, Perry R, Ireland T, Wills AK, Sandy J, Ness A (2017) Are people with an orofacial cleft at a higher risk of dental caries? A systematic review and meta-analy. Br Dent J 223(1):37–47. https://doi.org/10.1038/sj.bdj.2017.581
Bastos Lages EM, Marcos B, Pordeus IA (2004) Oral Health of Individuals with Cleft Lip, Cleft Palate, or Both. Cleft Palate-Craniofacial J 41(1):59–63. https://doi.org/10.1597/02-058
Allam GG, Sobeh IA (2020) Caries Experience Varies in Egyptian Children With Different Combinations of Cleft Lip and Palate and Is Related to Carbohydrate Intake Between Meals. Cleft Palate-Craniofacial J. https://doi.org/10.1177/1055665620952297
Mysak J, Podzimek S, Sommerova P et al (2014) Porphyromonas gingivalis: Major periodontopathic pathogen overview. J Immunol Res 2014. https://doi.org/10.1155/2014/476068
Lamont RJ, Koo H, Hajishengallis G (2018) The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol 16(12):745–759. https://doi.org/10.1038/s41579-018-0089-x
Mombelli A, Urs B, Niklaus L (1992) Microbiota associated with resiudal clefts and neighboring teeth in patients with cleft lip, alveolus and palate. Cleft Palate-Craniofacial J 29(5):463–469
Weckwerth PH, De Mattias Franco AT, De Magalhães Lopes CA et al (2014) Bacterial Pathogens Related to Chronic Suppurative Otitis Media in Individuals With Cleft Palate: Bacteriological Culture and Polymerase Chain Reaction. Cleft Palate-Craniofacial J 51(2):145–153. https://doi.org/10.1597/11-325
Veiga KA, Porto AN, Matos FZ et al (2017) Caries Experience and Periodontal Status in Children and Adolescents with Cleft Lip and Palate. Pediatr Dent 39(2):139–144
Plakwicz P, Wyrębek B, Górska R, Cudziło D (2017) Periodontal Indices and Status in 34 Growing Patients with Unilateral Cleft Lip and Palate: A Split-Mouth Study. Int J Periodontics Restorative Dent 37(6):e344–e353. https://doi.org/10.11607/prd.3461
Wyrębek B, Cudziło D, Plakwicz P (2017) Evaluation of periodontal tissues in growing patients with bilateral cleft lip and palate. A pilot study. Dev period Med 21(2):154–161
Adeyemo WL, Adeyemi MO, Ogunsola FT et al (2013) Prevalence and bacteriology of bacteremia associated with cleft lip and palate surgery. J Craniofac Surg 24(4):1126–1131. https://doi.org/10.1097/SCS.0b013e31828016e8
Chuo CB, Timmons MJ (2005) The bacteriology of children before primary cleft lip and palate surgery. Cleft Palate-Craniofacial J 42(3):272–276. https://doi.org/10.1597/03-108.1
Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, Leys EJ, Paster BJ (2008) Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol 46:1407–1417
Arweiler NB, Netuschil L (2016) The Oral Microbiota. Adv Exp Med Biol 902:45–60. https://doi.org/10.1007/978-3-319-31248-4_4
Baker JL, Mark Welch JL, Kauffman KM, McLean JS, He X (2024) The oral microbiome: diversity, biogeography and human health_ h. Nat Rev. Microbiol 22(2):89–104
Vartoukian SR, Adamowska A, Lawlor M, Moazzez R, Dewhirst FE, Wade WG (2016) In Vitro Cultivation of 'Unculturable' Oral Bacteria, Facilitated by Community Culture and Media Supplementation with Siderophores. PLoS One 11(1):e0146926. https://doi.org/10.1371/journal.pone.0146926
Balachandran M, Cross KL, Podar M (2020) Single-Cell Genomics and the Oral Microbiome. J Dent Res 99(6):613–620. https://doi.org/10.1177/0022034520907380
Rodicio Mdel R, Mendoza MC (2004) Identificación bacteriana mediante secuenciación de ARNr 16S: fundamento, metodología y aplicaciones en microbiología clínica [Identification of bacteria through 16S rRNA sequencing: principles, methods and applications in clinical microbiology]. Enferm Infecc Microbiol Clin 22(4):238–245. https://doi.org/10.1157/13059055
Ruparell A, Inui T, Staunton R, Wallis C, Deusch O, Holcombe LJ (2020) The canine oral microbiome: variation in bacterial populations across different niches. BMC Microbiol 20(1):42. https://doi.org/10.1186/s12866-020-1704-3
Funding
Open Access funding provided by Colombia Consortium No funding was obtained for this study.
Author information
Authors and Affiliations
Contributions
All authors wrote the main manuscript text TB, ME,FE prepared figures TB, ME,FE, DG prepared table FE,AS materials and methods MPB,FE,DG Discussion All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not Applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Escobar-Arregocés, F., Eras, MA., Bustos, A. et al. Characterization of the oral microbiota and the relationship of the oral microbiota with the dental and periodontal status in children and adolescents with nonsyndromic cleft lip and palate. Systematic literature review and meta-analysis. Clin Oral Invest 28, 245 (2024). https://doi.org/10.1007/s00784-024-05624-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00784-024-05624-3