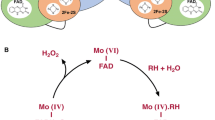
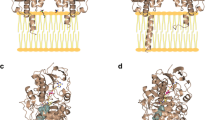
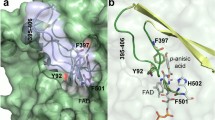
Abstract
In this work, a combination of homology modeling and molecular dynamics (MD) simulations was used to investigate the factors that modulate substrate specificity and activity of the mouse AOX isoforms: mAOX1, mAOX2 (previously mAOX3l1), mAOX3 and mAOX4. The results indicate that the AOX isoform structures are highly preserved and even more conserved than the corresponding amino acid sequences. The only differences are at the protein surface and substrate-binding site region. The substrate-binding site of all isoforms consists of two regions: the active site, which is highly conserved among all isoforms, and a isoform-specific region located above. We predict that mAOX1 accepts a broader range of substrates of different shape, size and nature relative to the other isoforms. In contrast, mAOX4 appears to accept a more restricted range of substrates. Its narrow and hydrophobic binding site indicates that it only accepts small hydrophobic substrates. Although mAOX2 and mAOX3 are very similar to each other, we propose the following pairs of overlapping substrate specificities: mAOX2/mAOX4 and mAOX3/mAXO1. Based on these considerations, we propose that the catalytic activity between all isoforms should be similar but the differences observed in the binding site might influence the substrate specificity of each enzyme. These results also suggest that the presence of several AOX isoforms in mouse allows them to oxidize more efficiently a wider range of substrates. This contrasts with the same or other organisms that only express one isoform and are less efficient or incapable of oxidizing the same type of substrates.






Similar content being viewed by others
References
Garattini E, Mendel R, Romão MJ, Wright R, Terao M (2003) Biochem J 372:15–32. doi:10.1042/BJ20030121
Garattini E, Fratelli M, Terao M (2009) Hum Genomics 4:119–130
Kurosaki M, Bolis M, Fratelli M, Barzago MM, Pattini L, Perretta G, Terao M, Garattini E (2013) Cell Mol Life Sci 70:1807–1830. doi:10.1007/s00018-012-1229-5
Terao M, Kurosaki M, Demontis S, Zanotta S, Garattini E (1998) Biochem J 332(Pt 2):383–393
Calzi ML, Raviolo C, Ghibaudi E, de Gioia L, Salmona M, Cazzaniga G, Kurosaki M, Terao M, Garattini E (1995) J Biol Chem 270:31037–31045
Garattini E, Fratelli M, Terao M (2008) Cell Mol Life Sci 65:1019–1048. doi:10.1007/s00018-007-7398-y
Kurosaki M, Terao M, Barzago MM, Bastone A, Bernardinello D, Salmona M, Garattini E (2004) J Biol Chem 279:50482–50498. doi:10.1074/jbc.M408734200
Garattini E, Terao M (2011) Drug Metab Rev 43:374–386. doi:10.3109/03602532.2011.560606
Garattini E, Terao M (2012) Expert Opin Drug Metab Toxicol 8:487–503. doi:10.1517/17425255.2012.663352
Garattini E, Terao M (2013) Expert Opin Drug Discov 8:641–654. doi:10.1517/17460441.2013.788497
Hartmann T, Terao M, Garattini E, Teutloff C, Alfaro JF, Jones JP, Leimkühler S (2012) Drug Metab Dispos 40:856–864. doi:10.1124/dmd.111.043828
Coelho C, Mahro M, Trincao J, Carvalho ATP, Ramos MJ, Terao M, Garattini E, Leimküehler S, Romão MJ (2012) J Biol Chem 287:40690–40702. doi:10.1074/jbc.M112.390419
Sali A, Blundell TL (1993) J Mol Biol 234:779–815. doi:10.1006/jmbi.1993.1626
Fiser A, Do RK, Sali A (2000) Protein Sci 9:1753–1773. doi:10.1110/ps.9.9.1753
Martí-Renom MA, Stuart AC, Fiser A, Sánchez R, Melo F, Sali A (2000) Annu Rev Biophys Biomol Struct 29:291–325. doi:10.1146/annurev.biophys.29.1.291
Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A (2006) Curr Protoc Bioinform. Chapter 5, Unit 5.6. doi:10.1002/0471250953.bi0506s15
Shen MY, Sali A (2006) Protein Sci 15:2507–2524. doi:10.1110/ps.062416606
Becke AD (1988) Phys Rev A 38:3098–3100
Becke AD (1996) J Chem Phys 104:1040–1046
Lee C, Yang W, Parr RG (1988) Phys Rev B Condens Matter 37:785–789
Neves RPP, Sousa SF, Fernandes PA, Ramos MJ (2013) J Chem Theory Comput 9:2718–2732
Bayly CI, Cieplak P, Cornell W, Kollman PA (1993) J Phys Chem 97:10269–10280
Batsanov SS (2001) Inorg Mater 37:871–885
Carvalho AT, Teixeira AF, Ramos MJ (2013) J Comput Chem 34:1540–1548. doi:10.1002/jcc.23287
Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Götz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh MJ, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA (2012) Amber 12. University of California, San Francisco
Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Proteins 65:712–725. doi:10.1002/prot.21123
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) J Comput Chem 25:1157–1174. doi:10.1002/jcc.20035
Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) J Am Chem Soc 117:5179–5197
Bras NFB, Cerqueira NMFSA, Fernandes PA, Ramos MJ (2008) Int J Quantum Chem 108:2030–2040
Gesto DS, Cerqueira NMFSA, Fernandes PA, Ramos MJ (2013) J Amer Chem Soc 135:7146–7158. doi:10.1021/ja310165u
Mahro M, Brás NF, Cerqueira NM, Teutloff C, Coelho C, Romão MJ, Leimkühler S (2013) PLoS One 8:e82285. doi:10.1371/journal.pone.0082285
Izaguirre JA, Catarello DP, Wozniak JM, Skeel RD (2001) J Chem Phys 114:2090–2098
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) J Comput Phys 23:327–341
Essman U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) J Chem Phys 103:8577–8593
Ribeiro JV, Tamames JA, Cerqueira NM, Fernandes PA, Ramos MJ (2013) Chem Biol Drug Des 82:743–755. doi:10.1111/cbdd.12197
Ribeiro JV, Cerqueira NMFSA, Moreira IS, Fernandes PA, Ramos MJ (2012) Theor Chem Acc 131:1271
Vila R, Kurosaki M, Barzago MM, Kolek M, Bastone A, Colombo L, Salmona M, Terao M, Garattini E (2004) J Biol Chem 279:8668–8683. doi:10.1074/jbc.M308137200
Acknowledgments
The authors would like to acknowledge the program FEDER/COMPETE and the Fundação para a Ciência e Tecnologia (FCT) for financial support [Projects PEst-C/EQB/LA0006/2013;EXCL/QEQ-COM/0394/2012; PTDC/BIA-PRO/118377/2010; and Grant SFRH/BPD/84581/2012 (CC)].
Author information
Authors and Affiliations
Corresponding authors
Additional information
N. M. F. S. A. Cerqueira, C. Coelho and N. F. Brás contributed equally to this work.
Responsible Editors: José Moura and Paul Bernhardt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
775_2014_1198_MOESM1_ESM.pdf
Figure S1 Sequence alignment of the mAOX1, mAOX3, mAOX2 and mAOX4 isoforms. Colored spheres mark the Isoenzyme-specific amino acid residues (mAOX1 in pink; mAOX3 in green; mAOX2 in orange; and mAOX4 in purple) (PDF 1155 kb)
Rights and permissions
About this article
Cite this article
Cerqueira, N.M.F.S.A., Coelho, C., Brás, N.F. et al. Insights into the structural determinants of substrate specificity and activity in mouse aldehyde oxidases. J Biol Inorg Chem 20, 209–217 (2015). https://doi.org/10.1007/s00775-014-1198-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-014-1198-2