Abstract
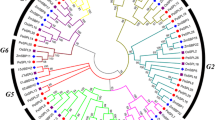
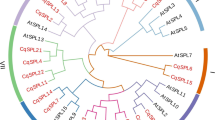
A transcription factor in plants encodes SQUAMOSA promoter binding protein-like (SPL) serves a broad spectrum of important roles for the plant, like, growth, flowering, and signal transduction. A gene family that encodes SPL proteins is documented in various model plant species, including Arabidopsis thaliana and Oryza sativa. Chickpea (Cicer arietinum), a leguminous crop, has not been thoroughly explored with regard to the SPL protein-encoding gene family. Chickpea SPL family genes were located and characterized computationally using a genomic database. Gene data of chickpea were obtained from the phytozome repository and was examined using bioinformatics methods. For investigating the possible roles of SPLs in chickpea, genome-wide characterization, expression, as well as structural analyses of this SPL gene family were performed. Cicer arietinum genome had 19 SPL genes, whereas, according to phylogenetic analysis, the SPLs in chickpea are segregated among four categories: Group-I has 2 introns, Group-II and IV have 1–2 introns (except CaSPL13 and CaSPL15 having 3 introns), and Group-III has 9 introns (except CaSPL1 and CaSPL11 with 1 and 8 introns, respectively). The SBP domain revealed that SPL proteins featured two zinc-binding sites, i.e., C3H and C2HC and one nuclear localization signal. All CaSPL proteins are found to contain highly conserved motifs, i.e., Motifs 1, 2, and 4, except CaSPL10 in which Motifs 1 and 4 were absent. Following analysis, it was found that Motifs 1 and 2 of the chickpea SBP domain are Zinc finger motifs, and Motif 4 includes a nuclear localization signal. All pairs of CaSPL paralogs developed by purifying selection. The CaSPL promoter investigation discovered cis-elements that are responsive to stress, light, and phytohormones. Examination of their expression patterns highlighted major CaSPLs to be evinced primarily among younger pods and flowers. Indicating their involvement in the plant’s growth as well as development, along with their capacity to react as per different situations by handling the regulation of target gene’s expression, several CaSPL genes are also expressed under certain stress conditions, namely, cold, salt, and drought. The majority of the CaSPL genes are widely expressed and play crucial roles in terms of the plant’s growth, development, and responses to the environmental-stress conditions. Our work provides extensive insight into the gene family CaSPL, which might facilitate further studies related to the evolution and functions of the SPL genes for chickpea and other plant species.










Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- CaSPL:
-
Chickpea SPL
- CDS:
-
Coding sequence
- HMM:
-
Hidden Markov model
- SPL:
-
SQUAMOSA promoter-binding protein-like
- NLS:
-
Nuclear localization signal
- SMART:
-
Simple Modular Architecture Research Tool
- GRAVY:
-
Grand average of hydropathicity
- pI:
-
Isoelectric point
- MW:
-
Molecular weight
- MEME:
-
Multiple Em for Motif Elicitation
- MCScanX:
-
Multiple Collinearity Scan toolbox
- PPI:
-
Protein-protein interaction
- TF:
-
Transcription factor
References
Acharjee S, Sarmah BK (2013) Biotechnologically generating “super chickpea” for food and nutritional security. Plant Sci 207:108–116. https://doi.org/10.1016/J.PLANTSCI.2013.02.003
Altschul SF, Madden TL, Schäffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402. https://doi.org/10.1093/nar/25.17.3389
Bai F, Reinheimer R, Durantini D et al (2012) TCP transcription factor, BRANCH ANGLE DEFECTIVE 1 (BAD1), is required for normal tassel branch angle formation in maize. Proc Natl Acad Sci U S A 109:12225–12230. https://doi.org/10.1073/PNAS.1202439109
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME suite. Nucleic Acids Res 43:W39–W49. https://doi.org/10.1093/NAR/GKV416
Bioinformatics B, Valencia S (2019) OmicsBox—Bioinformatics made easy (Version 2.1.14). https://www.biobam.com/omicsbox
Birkenbihl RP, Jach G, Saedler H, Huijser P (2005) Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol 352:585–596. https://doi.org/10.1016/J.JMB.2005.07.013
Blum M, Chang HY, Chuguransky S et al (2021) The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49:D344–D354. https://doi.org/10.1093/NAR/GKAA977
Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438. https://doi.org/10.1038/NATURE01521
Cannon SB, Mitra A, Baumgarten A et al (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:1–21. https://doi.org/10.1186/1471-2229-4-10
Cardon GH, Höhmann S, Nettesheim K et al (1997) Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J 12:367–377. https://doi.org/10.1046/J.1365-313X.1997.12020367.X
Chen C, Chen H, Zhang Y et al (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202. https://doi.org/10.1016/J.MOLP.2020.06.009
Cheng H, Hao M, Wang W et al (2016) Genomic identification, characterization and differential expression analysis of SBP-box gene family in Brassica napus. BMC Plant Biol 16:1–17. https://doi.org/10.1186/S12870-016-0852-Y
Chou KC, Shen HB (2010) Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 5(6):e11335. https://doi.org/10.1371/journal.pone.0011335
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39:W155–W159. https://doi.org/10.1093/NAR/GKR319
Eddy SR (2009) A new generation of homology search tools based on probabilistic inference. Genome informatics. International Conference on Genome Informatics 23(1):205–211
Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17:3470–3488. https://doi.org/10.1105/tpc.105.035659
Gao J, Ge W, Zhang Y et al (2015) Identification and characterization of microRNAs at different flowering developmental stages in moso bamboo (Phyllostachys edulis) by high-throughput sequencing. Mol Genet Genomics 290:2335–2353. https://doi.org/10.1007/S00438-015-1069-8
Gao J, Peng H, Chen F, Liu Y, Chen B et al (2019) Genome-wide identification and characterization, phylogenetic comparison and expression profiles of SPL transcription factor family in B. juncea (Cruciferae). PLos One 14(11):e0224704. https://doi.org/10.1371/journal.pone.0224704
Gasteiger E, Hoogland C, Gattiker A et al (2005) Protein identification and analysis tools on the ExPASy server. The Proteomics Protocols Handbook 571–607. https://doi.org/10.1385/1-59259-890-0:571
Gielen H, Remans T, Vangronsveld J, Cuypers A (2016) Toxicity responses of Cu and Cd: the involvement of miRNAs and the transcription factor SPL7. BMC Plant Biol 16:1–16. https://doi.org/10.1186/S12870-016-0830-4
Gu Z, Steinmetz LM, Gu X et al (2003) Role of duplicate genes in genetic robustness against null mutations. Nature 2002 421:6918 421:63–66. https://doi.org/10.1038/nature01198
Han YY, Ma YQ, Li DZ et al (2016) Characterization and phylogenetic analysis of fifteen NtabSPL genes in Nicotiana tabacum L. cv. Qinyan95. Dev Genes Evol 226:1–14. https://doi.org/10.1007/S00427-015-0522-3
Horton P, Park KJ, Obayashi T et al (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:39–48. https://doi.org/10.1093/nar/gkm259
Hou H, Li J, Gao M et al (2013) Genomic organization, phylogenetic comparison and differential expression of the SBP-box family genes in grape. PLoS ONE 8:e59358. https://doi.org/10.1371/JOURNAL.PONE.0059358
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene features visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
İlhan E, Büyük İ, İnal B (2018) Transcriptome — scale characterization of salt responsive bean TCP transcription factors. Gene 642:64–73. https://doi.org/10.1016/J.GENE.2017.11.021
Ishii T, Numaguchi K, Miura K et al (2013) OsLG1 regulates a closed panicle trait in domesticated rice. Nat Genet 45:462–465. https://doi.org/10.1038/NG.2567
Jin J, Tian F, Yang DC et al (2017) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45:D1040–D1045. https://doi.org/10.1093/NAR/GKW982
Jung JH, Lee HJ, Ryu JY, Park CM (2016) SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol Plant 9:1647–1659. https://doi.org/10.1016/J.MOLP.2016.10.014
Klein J, Saedler H, Huijser P (1996) A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol Gen Genet 250:7–16. https://doi.org/10.1007/BF02191820
Kropat J, Tottey S, Birkenbihl RP et al (2005) A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc Natl Acad Sci U S A 102:18730. https://doi.org/10.1073/PNAS.0507693102
Lee J, Park JJ, Kim SL et al (2007) Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol Biol 65:487–499. https://doi.org/10.1007/S11103-007-9196-1
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327. https://doi.org/10.1093/nar/30.1.325
Letunic I, Khedkar S, Bork P (2021) SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49:D458–D460. https://doi.org/10.1093/NAR/GKAA937
Li C, Lu S (2014) Molecular characterization of the SPL gene family in Populus trichocarpa. BMC Plant Biol 14:1–15. https://doi.org/10.1186/1471-2229-14-131
Li J, Hou H, Li X et al (2013) Genome-wide identification and analysis of the SBP-box family genes in apple (Malus × domestica Borkh.). Plant Physiol Biochem 70:100–114. https://doi.org/10.1016/J.PLAPHY.2013.05.021
Liang X, Nazarenus TJ, Stone JM (2008) Identification of a consensus DNA-binding site for the Arabidopsis thaliana SBP domain transcription factor, AtSPL14, and binding kinetics by surface plasmon resonance. Biochemistry 47:3645–3653. https://doi.org/10.1021/BI701431Y/SUPPL_FILE/BI701431Y_SI.PDF
Manning K, Tör M, Poole M et al (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38:948–952. https://doi.org/10.1038/NG1841
Mistry J, Chuguransky S, Williams L et al (2021) Pfam: the protein families database in 2021. Nucleic Acids Res 49:D412–D419. https://doi.org/10.1093/NAR/GKAA913
Miura K, Ikeda M, Matsubara A et al (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42:545–549. https://doi.org/10.1038/ng.592
Moreno MA, Harper LC, Krueger RW et al (1997) liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev 11:616–628. https://doi.org/10.1101/GAD.11.5.616
Muehlbauer FJ, Sarker A (2017) Economic importance of chickpea: production, value, and world trade. In: Varshney R, Thudi M, Muehlbauer F (eds) The Chickpea Genome. Compendium of Plant Genomes. Springer, Cham. https://doi.org/10.1007/978-3-319-66117-9_2
Oliver T, Schmidt B, Nathan D et al (2005) Using reconfigurable hardware to accelerate multiple sequence alignment with ClustalW. Bioinformatics 21:3431–3432. https://doi.org/10.1093/BIOINFORMATICS/BTI508
Padmanabhan MS, Ma S, Burch-Smith TM et al (2013) Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog 9:e1003235. https://doi.org/10.1371/JOURNAL.PPAT.1003235
Pan F, Wang Y, Liu H et al (2017) Genome-wide identification and expression analysis of SBP-like transcription factor genes in moso bamboo (Phyllostachys edulis). BMC Genomics 18:1–17. https://doi.org/10.1186/S12864-017-3882-4
Preston JC, Hileman LC (2013) Functional evolution in the plant SQUAMOSA-promoter binding protein-like (SPL) gene family. Front Plant Sci 4:80–80. https://doi.org/10.3389/FPLS.2013.00080
Rouster J, Leah R, Mundy J, Cameron-Mills V (1997) Identification of a methyl jasmonate-responsive region in the promoter of a lipoxygenase 1 gene expressed in barley grain. Plant J Cell Mol Biol 11:513–523. https://doi.org/10.1046/j.1365-313x.1997.11030513.x
Salinas M, Xing S, Höhmann S et al (2012) Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 235:1171–1184. https://doi.org/10.1007/S00425-011-1565-Y
Sani SGAS, Chang PL, Zubair A et al (2018) Genetic diversity, population structure, and genetic correlation with climatic variation in chickpea (Cicer arietinum) landraces from Pakistan. Plant Genome 11:170067. https://doi.org/10.3835/PLANTGENOME2017.08.0067
Shalom L, Shlizerman L, Zur N et al (2015) Molecular characterization of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) gene family from Citrus and the effect of fruit load on their expression. Front Plant Sci 6:1–10. https://doi.org/10.3389/FPLS.2015.00389
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/GR.1239303
Shiu SH, Karlowski WM, Pan R et al (2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16:1220–1234. https://doi.org/10.1105/TPC.020834
Silva GFFE, Silva EM, da Silva AM et al (2014) microRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J 78:604–618. https://doi.org/10.1111/TPJ.12493
Song A, Gao T, Wu D et al (2016) Transcriptome-wide identification and expression analysis of chrysanthemum SBP-like transcription factors. Plant Physiol Biochem 102:10–16. https://doi.org/10.1016/J.PLAPHY.2016.02.009
Stone JM, Liang X, Nekl ER, Stiers JJ (2005) Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J 41:744–754. https://doi.org/10.1111/J.1365-313X.2005.02334.X
Su R, Yuan B, Yang Y, Ao G, Wang J (2023) Genome-wide analysis of SPL gene families illuminate the evolution patterns in three rubber-producing plants. Diversity 15(9):983. https://doi.org/10.3390/d15090983
Szklarczyk D, Gable AL, Lyon D et al (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. https://doi.org/10.1093/NAR/GKY1131
Tamura K, Stecher G, Kumar S (2021) MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/MOLBEV/MSAB120
Thudi M, Khan AW, Kumar V et al (2016) Whole genome re-sequencing reveals genome-wide variations among parental lines of 16 mapping populations in chickpea (Cicer arietinum L.). BMC Plant Biol 16:53–64. https://doi.org/10.1186/S12870-015-0690-3/TABLES/3
Trapnell C, Roberts A, Goff L et al (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. https://doi.org/10.1038/NPROT.2012.016
Tripathi RK, Goel R, Kumari S, Dahuja A (2017) Genomic organization, phylogenetic comparison, and expression profiles of the SPL family genes and their regulation in soybean. Dev Genes Evol 227:101–119. https://doi.org/10.1007/S00427-017-0574-7
Unte US, Sorensen AM, Pesaresi P et al (2003) SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 15:1009–1019. https://doi.org/10.1105/TPC.010678
Varshney RK, Mohan SM, Gaur PM et al (2013) Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotechnol Adv 31:1120–1134. https://doi.org/10.1016/J.BIOTECHADV.2013.01.001
Wang H, Wang H (2015) The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol Plant 8:677–688. https://doi.org/10.1016/J.MOLP.2015.01.008
Wang H, Nussbaum-Wagler T, Li B et al (2005) The origin of the naked grains of maize. Nature 436:714–719. https://doi.org/10.1038/NATURE03863
Wang S, Wu K, Yuan Q et al (2012a) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44:950–954. https://doi.org/10.1038/NG.2327
Wang Z, Wang Y, Kohalmi SE et al (2016) SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 2 controls floral organ development and plant fertility by activating ASYMMETRIC LEAVES 2 in Arabidopsis thaliana. Plant Mol Biol 92:661–674. https://doi.org/10.1007/S11103-016-0536-X
Waqas M, Azhar MT, Rana IA et al (2019) Genome-wide identification and expression analyses of WRKY transcription factor family members from chickpea (Cicer arietinum L.) reveal their role in abiotic stress-responses. Genes Genom 41:467–481. https://doi.org/10.1007/s13258-018-00780-9
Xie K, Wu C, Xiong L (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol 142:280–293. https://doi.org/10.1104/PP.106.084475
Xing S, Salinas M, Höhmann S et al (2010) miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22:3935–3950. https://doi.org/10.1105/TPC.110.079343
Xing S, Salinas M, Garcia-Molina A et al (2013) SPL8 and miR156-targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. Plant J 75:566–577. https://doi.org/10.1111/TPJ.12221
Xu M, Hu T, Zhao J et al (2016) Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet 12:e1006263. https://doi.org/10.1371/JOURNAL.PGEN.1006263
Yadav S, Yadava YK, Meena S et al (2023) The SPL transcription factor genes are potential targets for epigenetic regulation in response to drought stress in chickpea (C. arietinum L.). Mol Biol Rep 50:5509–5517. https://doi.org/10.1007/s11033-023-08347-y
Yamaguchi A, Wu MF, Yang L et al (2009) The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell 17:268–278. https://doi.org/10.1016/J.DEVCEL.2009.06.007
Yamasaki K, Kigawa T, Inoue M et al (2004) A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J Mol Biol 337:49–63. https://doi.org/10.1016/J.JMB.2004.01.015
Yamasaki H, Hayashi M, Fukazawa M et al (2009) SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21:347–361. https://doi.org/10.1105/tpc.108.060137
Yang Z, Wang X, Gu S et al (2008) Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 407:1–11. https://doi.org/10.1016/J.GENE.2007.02.034
Yin J, Liu M, Ma D et al (2018) Identification of circular RNAs and their targets during tomato fruit ripening. Postharvest Biol Technol 136:90–98. https://doi.org/10.1016/J.POSTHARVBIO.2017.10.013
Zhang SD, Ling LZ (2014) Genome-wide identification and evolutionary analysis of the SBP-box gene family in castor bean. PLoS ONE 9:e86688. https://doi.org/10.1371/JOURNAL.PONE.0086688
Zhang Y, Schwarz S, Saedler H, Huijser P (2006) SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol Biol 63:429–439. https://doi.org/10.1007/S11103-006-9099-6
Zhang X, Dou L, Pang C et al (2014) Genomic organization, differential expression, and functional analysis of the SPL gene family in Gossypium hirsutum. Mol Genet Genom 290(1):115–126. https://doi.org/10.1007/S00438-014-0901-X
Zhang HX, Jin JH, He YM et al (2016) Genome-wide identification and analysis of the SBP-box family genes under Phytophthora capsici stress in pepper (Capsicum annuum L.). Front Plant Sci 7:504. https://doi.org/10.3389/FPLS.2016.00504
Zhao H, Cao H, Zhang M, Deng S, Li T, Xing S (2022) Genome-wide identification and characterization of SPL family genes in Chenopodium quinoa. Genes (Basel) 13(8):1455. https://doi.org/10.3390/genes13081455
Zhu Z, Tan L, Fu Y et al (2013) Genetic control of inflorescence architecture during rice domestication. Nat Commun 4:1–8. https://doi.org/10.1038/NCOMMS3200
Zhu YX, Yang L, Liu N et al (2019) Genome-wide identification, structure characterization, and expression pattern profiling of aquaporin gene family in cucumber. BMC Plant Biol 19:345. https://doi.org/10.1186/S12870-019-1953-1
Acknowledgements
The authors acknowledge the Department of Biotechnology and Microbiology, School of Sciences, Noida International University, Gautam Budh Nagar, U.P., India for the required facilities.
Funding
A. P. thanks the Department of Biotechnology, Ministry of Science and Technology, Govt. of India for MK Bhan Young Researcher Fellowship Program (HRD-12/4/2020-AFS-DBT-Part (1) (13815).
Author information
Authors and Affiliations
Contributions
S. S. conceived and designed the research. S. S. conducted the experiments. S. S. and A. P. contributed the analytical tools. S. S. and P. B. analyzed the data. S. S. wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Handling Editor: Bhumi Nath Tripathi
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, S., Praveen, A. & Bhadrecha, P. Genome-wide identification and analysis of SPL gene family in chickpea (Cicer arietinum L.). Protoplasma (2024). https://doi.org/10.1007/s00709-024-01936-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00709-024-01936-z