Abstract
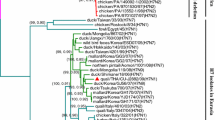
Genotype 4 (G4) Eurasian avian-like lineage swine H1N1 influenza A viruses, which are reassortants containing sequences from the pandemic 2009 H1N1 virus lineage, triple-reassortant-lineage internal genes, and EA-lineage external genes, have been reported in China since 2013. These have been predominant in pig populations since 2016 and have exhibited pandemic potential. In this study, we developed a one-step multiplex RT-qPCR assay targeting the M, HA1, and PB2 genes to detect G4 and related EA H1N1 viruses, with detection limits of 1.5 × 101 copies/μL and 1.15 × 10−2 ng/μL for the purified PCR products and RNA templates, respectively. The specificity of the detection method was confirmed using various influenza virus subtypes. When the one-step multiplex RT-qPCR assay was applied to swine respiratory samples collected between 2020 and 2022 in Korea, a virus related to G4 EA H1N1 strains was detected. Phylogenetic analysis based on portions of all eight genome segments showed that the positive sample contained HA, NA, PB2, NS, and NP genes closely related to those of G4 EA H1N1 viruses, confirming the ability of our assay to accurately detect G4 EA H1N1 viruses in the field.

Similar content being viewed by others
Data availability
The sequences generated in this study were deposited in the GenBank database under the accession numbers listed in Supplementary Table S1.
References
Anderson TK, Macken CA, Lewis NS et al (2016) A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza A viruses. mSphere 1(6):e00275-00216
Belshe RB (2005) The origins of pandemic influenza–lessons from the 1918 virus. N Engl J Med 353(21):2209–2211
Chang CY, Deng MC, Wang FI et al (2014) The application of a duplex reverse transcription real-time PCR for the surveillance of porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. J Virol Methods 201:13–19
Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team, Dawood FS, Jain S et al (2009) Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 360(25):2605–2615
Freidl GS, Meijer A, de Bruin E et al (2014) Influenza at the animal-human interface: a review of the literature for virological evidence of human infection with swine or avian influenza viruses other than A(H5N1). Euro Surveill 19(18):1
Gu M, Chen K, Ge Z et al (2022) Zoonotic threat of G4 genotype Eurasian avian-like swine influenza A(H1N1) Viruses, China, 2020. Emerg Infect Dis 28(8):1664–1668
Guo F, Yang J, Pan J et al (2020) Origin and evolution of H1N1/pdm2009: A codon usage perspective. Front Microbiol 11:1615
Hoffmann E, Neumann G, Kawaoka Y et al (2000) A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA 97(11):6108–6113
Huang YL, Pang VF, Pan CH et al (2009) Development of a reverse transcription multiplex real-time PCR for the detection and genotyping of classical swine fever virus. J Virol Methods 160(1–2):111–118
Javanian M, Barary M, Ghebrehewet S et al (2021) A brief review of influenza virus infection. J Med Virol 93(8):4638–4646
Krammer F, Smith GJD, Fouchier RAM et al (2018) Influenza. Nat Rev Dis Primers 4(1):3
Le TB, Kim HK, Na W et al (2020) Development of a multiplex RT-qPCR for the detection of different clades of avian influenza in poultry. Viruses 12(1):1
Li X, Guo L, Liu C et al (2019) Human infection with a novel reassortant Eurasian-avian lineage swine H1N1 virus in northern China. Emerg. Microbes Infect. 8(1):1535–1545
Li Z, Zhao X, Huang W et al (2022) Etiological characteristics of the first human infection with the G4 genotype Eurasian avian⁃ like H1N1 swine influenza virus in Yunnan Province China. Chin. J. Virol. 38:290–297
Long JS, Mistry B, Haslam SM et al (2019) Host and viral determinants of influenza A virus species specificity. Nat Rev Microbiol 17(2):67–81
Ma W, Kahn RE, Richt JA (2008) The pig as a mixing vessel for influenza viruses: human and veterinary implications. J Mol Genet Med 3(1):158–166
Meng F, Chen Y, Song Z et al (2022) Continued evolution of the Eurasian avian-like H1N1 swine influenza viruses in China. Sci. China Life Sci. 1:1
Meng F, Yang H, Qu Z et al (2022) A Eurasian avian-like H1N1 swine influenza reassortant virus became pathogenic and highly transmissible due to mutations in its PA gene. Proc Natl Acad Sci USA 119(34):e2203919119
Mondiale de la Santé O (2022) World Health Organization 2022 Antigenic and genetic characteristics of zoonotic influenza A viruses and development of candidate vaccine viruses for pandemic preparedness–Caractéristiques génétiques et antigéniques des virus grippaux A zoonotiques et mise au point de virus vaccinaux candidats pour se préparer à une pandémie. Week Epidemiol Record 97:120–132
Parys A, Vandoorn E, King J et al (2021) Human infection with Eurasian avian-like swine influenza A(H1N1) Virus, the Netherlands, September 2019. Emerg Infect Dis 27(3):939–943
Pensaert M, Ottis K, Vandeputte J et al (1981) Evidence for the natural transmission of influenza A virus from wild ducks to swine and its potential importance for man. Bull World Health Organ 59(1):75–78
Spackman E, Senne DA, Myers TJ et al (2002) Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol 40(9):3256–3260
Steel J, Lowen AC (2014) Influenza A virus reassortment. Curr Top Microbiol Immunol 385:377–401
Sun H, Xiao Y, Liu J et al (2020) Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc Natl Acad Sci USA 117(29):17204–17210
Sun H, Liu J, Xiao Y et al (2021) Pathogenicity of novel reassortant Eurasian avian-like H1N1 influenza virus in pigs. Virology 561:28–35
Suzuki T, Horiike G, Yamazaki Y et al (1997) Swine influenza virus strains recognize sialylsugar chains containing the molecular species of sialic acid predominantly present in the swine tracheal epithelium. FEBS Lett 404(2–3):192–196
Taubenberger JK, Morens DM (2006) 1918 Influenza: the mother of all pandemics. Emerg Infect Dis 12(1):15–22
Vijaykrishna D, Smith GJ, Pybus OG et al (2011) Long-term evolution and transmission dynamics of swine influenza A virus. Nature 473(7348):519–522
Wang SY, Wen F, Yu LX et al (2022) Potential threats to human health from Eurasian avian-like swine influenza A(H1N1) virus and its reassortants. Emerg Infect Dis 28(7):1489–1493
Acknowledgements
This work was supported by the Bio & Medical Technology Development Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science & ICT (2021M3E5E3083402), and the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2020R1A6A1A06046235).
Funding
This article was funded by Ministry of Science and ICT, South Korea (2021M3E5E3083402), and Ministry of Education (2020R1A6A1A06046235).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Handling Editor: William G Dundon.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Moon, S.H., Na, W., Shin, S. et al. Evidence for the circulation of genotype 4 Eurasian avian-like lineage swine H1N1 influenza A viruses on Korean swine farms, obtained using a newly developed one-step multiplex RT-qPCR assay. Arch Virol 168, 267 (2023). https://doi.org/10.1007/s00705-023-05887-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-023-05887-3